Projects Imaging Mass Spectrometry
Ongoing projects
- Can a smart surgical knife help brain surgeons remove tumours?
- High throughout spectral imaging with pixelated detectors
- Polyimage: Innovations in ambient imaging mass spectrometry
- Imaging interfaces in geochemistry
- Brainpath – Molecular Imaging of Brain Pathophysiology
- Imaging hypoxia and tumor heterogeneity
- TargetCare: New approaches to joint diseases
- PRISAR: Innovation in intra-operative diagnostics
- Imaging Mass Spectrometry in 3D digital pathology
- Intra-operative diagnostics for oncological procedures
- Metabolic profiling of bone during trauma surgery
- Molecular histology with imaging MS for colorectal cancer studies
- Molecular imaging of osteoarthritic cartilage and regeneration
- Intra-tumor heterogeneity and personalized medicine
- Unraveling the complexity of troponins in human heart by imaging mass spectrometry
- Forensic hair testing
- Quantitative molecular imaging in pharmaceutical sciences
- Project Eurlipids - Interreg EMR
Can a smart surgical knife help brain surgeons remove tumours?
[Click here for the original story in Dutch]
When removing a tumour, it is not always easy for a surgeon to be sure that all cancer cells have been removed. Removing too much healthy tissue is obviously not desirable, and in fact, sparing organ tissue is vital. Especially when it comes to the brain.
Scientists at the Maastricht MultiModal Molecular Institute (M4I) developed their ‘intelligent knife’, or iKnife, in recent years. In mutual collaboration between M4i and UZ Leuven, a very extensive study of the knife's clinical application has been carried out in recent years. This showed that the iKnife can recognise tissue of brain tumours, among others, within seconds while cutting, with more than 98% precision. The European Interreg Flanders-Netherlands grant programme has made over EUR 2 million available for the further development of this technique.
Molecular brain navigator tool
The iKnife is an electrosurgical tool that generates smoke when cutting tissue during surgery. This smoke contains molecules that act as a kind of fingerprint, revealing key information about the tissue. By analyzing this molecular fingerprint, surgeons can differentiate a tumour from the surrounding tissue—information that is invisible to the naked eye. However, the size of the original analysis device posed challenges in the operating room during earlier tests. To address this, the research team is now developing a more user-friendly handheld screener specifically designed for brain surgeons.
"Making our current iKnife, or molecular navigator, more compact is challenging because we aim to maintain both the sensitivity and accuracy of the existing system," explains M4I researcher Eva Cuypers. "This presents several technical hurdles. For example, we need to use a different type of detector to achieve a compact design. This also means developing a smaller component for the smoke supply. However, the underlying principle will remain the same, allowing us to compare molecular profiles from the new system with those from our previous model. Artificial Intelligence will help us determine if we can successfully translate our current model to this new system."
Brain Tumours
The ‘intelligent surgical knife’ uses mass spectrometry, a special analysis technique that can significantly improve surgical outcomes. Currently, around 60% of brain tumour patients experience a relapse within five years, as residual tumour cells often remain after surgery. How could the more precise tissue recognition offered by the iKnife benefit individual brain tumour patients?
"The iKnife will likely enable quicker identification of tumour cells during brain surgery," explains Maastricht neurosurgeon Olaf Schijns. "By analysing the molecules in the smoke generated by the iKnife, we can remove brain tumours faster and more thoroughly, without damaging ‘healthy’ tissue. Once we have sufficient evidence of its benefits for brain tumour patients, it’s even possible we could use this technique for other patient groups, such as those with severe epilepsy."
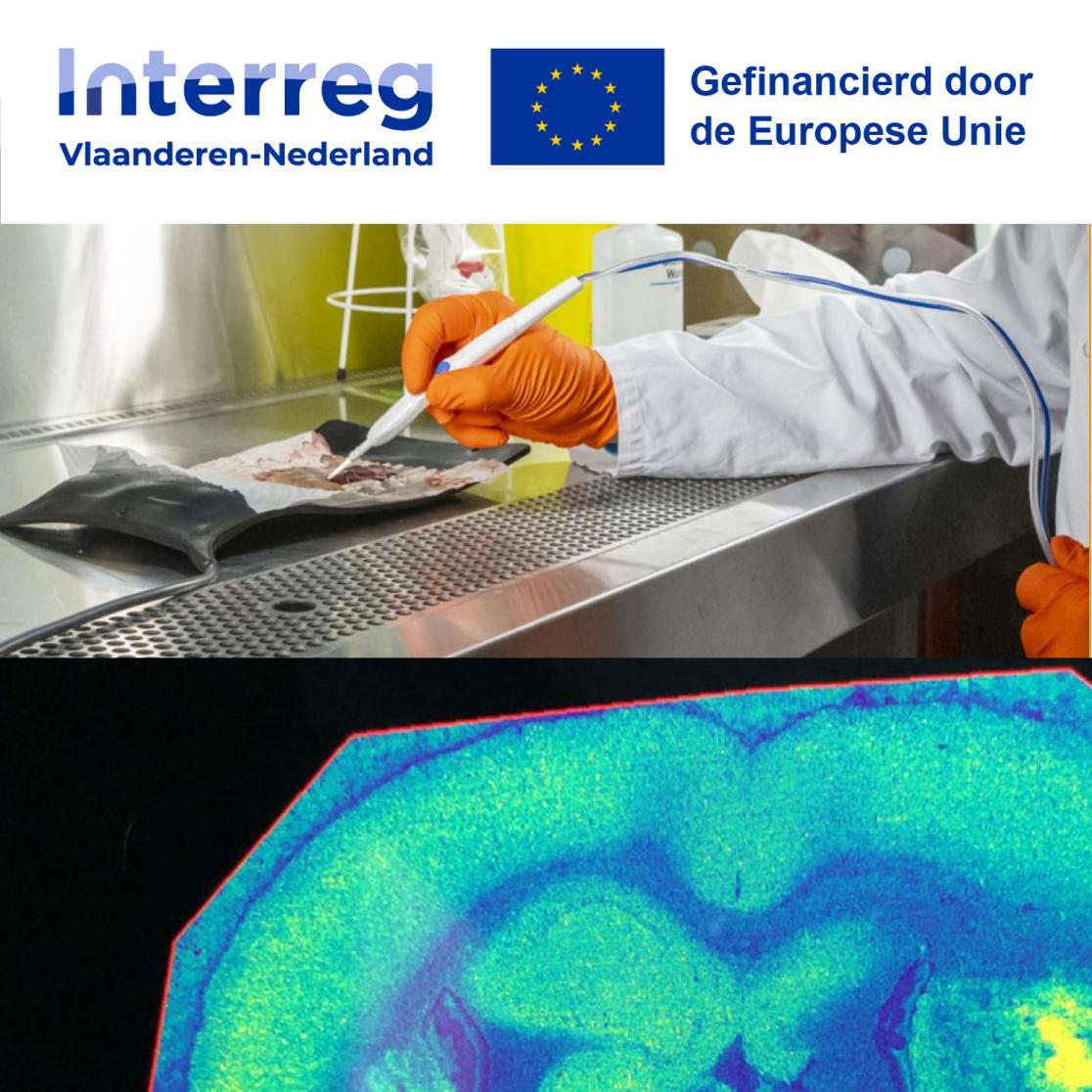
To ensure the successful development of the molecular navigator, a collaborative team of researchers, surgeons and companies has been assembled thanks to the support of Interreg Flanders-Netherlands, the Province of Limburg and Dutch national co-financing companies like Next Generation Sensors (for co-developing the compact navigator) and Aspect Analytics (for statistical analysis and a user-friendly interface). Neurosurgeons from Maastricht UMC+, Zuyderland in Sittard, UZ Leuven, Ziekenhuis Oost-Limburg (Genk), and AZ Groeninge (Kortrijk) will provide input and test the developed brain navigator as end-users throughout the development process.
High throughout spectral imaging
with pixelated detectors
This STW project aims to develop new direct imaging detector systems based on the Medipix/Timepix chip for applications in X-ray, ion and electron imaging. The focus areas in biomedical imaging are molecular histology with mass spectrometry and low-dose, energy dispersive X-ray imaging. We target the development of an ion imaging system that is suitable for the detection of macromolecular ions of either polarity, which can be post-accelerated up to 15kV. In this way, we create a bio-molecular imaging device for molecular histology for use with ion mobility and time-of-flight mass spectrometers. A complimentary imaging approach aims at energy dispersive X-ray imaging. A novel system will be developed with a new MPX chip based device that makes it possible to separate the incoming X-rays based on their energy and use eight thresholds to make seven windows in which one can count the incoming photons. Our overall goal is to improve quality and reduce the measuring time in ion and electron imaging and introduce energy information into X-ray imaging and thereby reduce the required dose.
Microscope mode mass spectrometry imaging using a pixelated detector
Bare chips of the Timepix type in combination with microchannel plates (MCPs) are implemented on a triple focusing time-of-flight (TRIFT) mass spectrometer as a detector for microscope mode mass spectrometry imaging of organic molecules in biological samples at high speed. (Figure 1) Applying a high potential on the Timepix/MCP detector, a wide mass range and an increased signal-to-noise ratio can be achieved compared with other microscope mode instruments. [1-6]
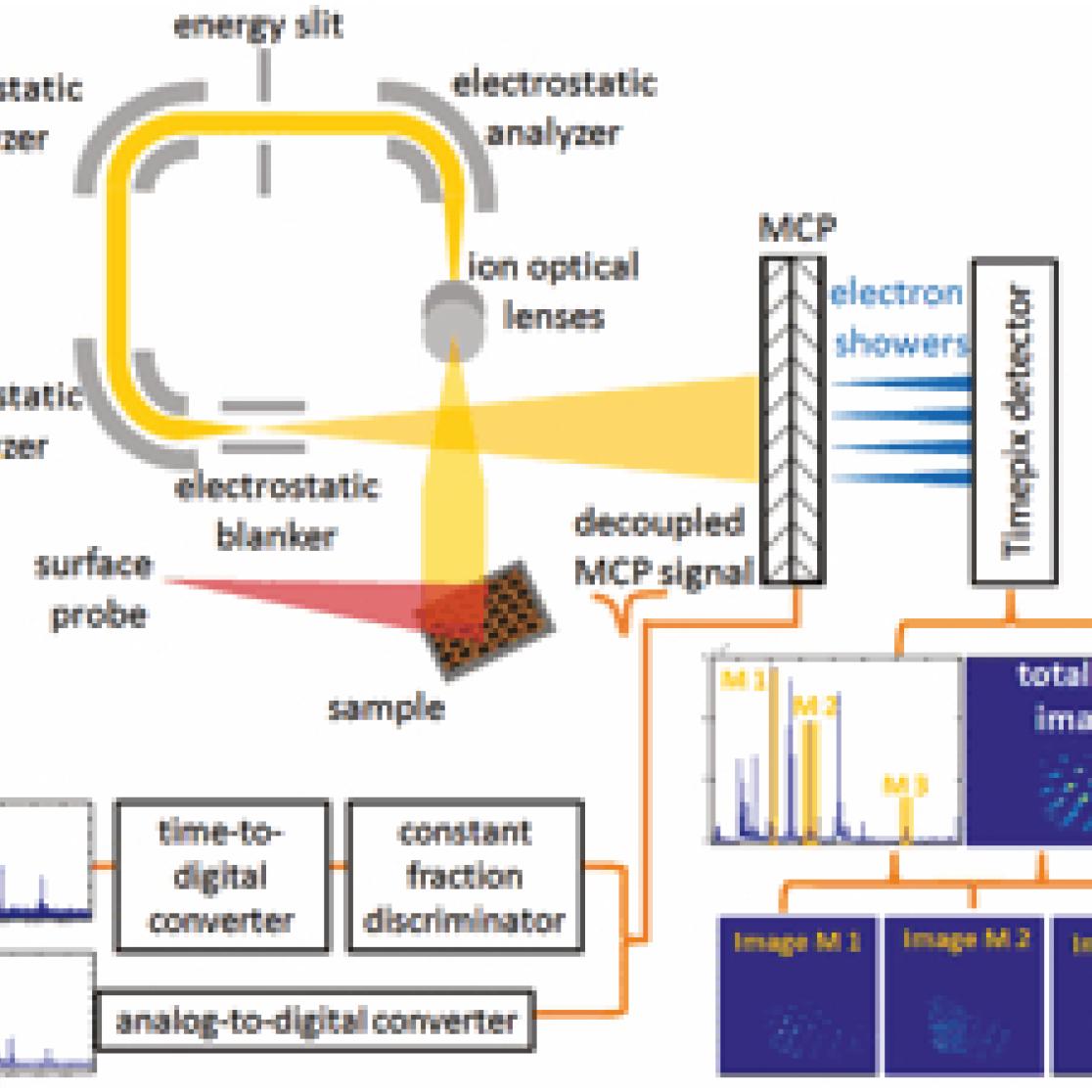
Figure 1 – Schematic overview of SIMS-trift ion microscope setup with implemented MCP/Timepix detector. Mass spectra are acquired without the need of mass selection. The spatial distribution of compounds on the sample surface are maintained during ionization and mass seperation and projected on the Timepix detector as such.
Polyimage: Innovations in ambient
imaging mass spectrometry
PolyImage - The development of a new chemical microscope for research of polymers and smart materials. Partners: Wageningen University, Maastricht University, DSM Resolve, Waters Chromatography, Omics2Image and RIKILT.
MSI is a technique in which molecules are locally extracted from a surface and ionized (electrically charged) after which their mass can be determined. The mass is then used to determine the molecular structure, which provides us with surface images of molecular distributions on top of the structural information provided by e.g. optical microscopes. This way the distribution of drug molecules and metabolites can be followed in tissue without labelling, or molecular surface changes to synthetic polymers can be monitored, for example.
Currently, MSI is not able to provide all molecular information from every type of sample: Many samples of interest, such as unfrozen tissue, unprocessed food or water containing polymers, are not compatible with high vacuum, which is needed for most current MSI methods as it provides much higher sensitivity. Liquid containing samples cannot be introduced into high-vacuum as the liquid would immediately evaporate. These samples are of high interest to both science and industry, for example to explain disease processes or improve food quality.
Molecules relevant for investigation, such as polymer additives or enzymes, are often present in relatively low abundancy, which makes the detection of these molecules much less likely. To be able to detect all molecules, the ionization method needs to be able to charge molecules along the full range of physical properties. For example, molecules with low proton affinities are not ionized by electrospray (attachment of a proton), which is the most efficient and common way of ionizing molecules at atmospheric pressure. In contrast, plasma ionization (detachment of an electron) is selective to molecules with low proton affinities.
Technical and chemical improvements
We aim to devise an instrument able to apply MSI on liquid containing samples with high sensitivity while also having the possibility to detect molecules with low proton affinities. Technical and chemical improvements in mass spectrometry imaging instruments must enable researchers to study the surface of (bio)polymers and smart materials more precisely. This new instrument will be more sensitive and applicable for measurement of chemicals on end products and intermediates, for trouble-shooting of products and production processes, for the development of new natural plastics, for the management of conversion of biomass and for the improvement of biosensors.
Imaging interfaces in geochemistry
Partner: Shell
In collaboration with Shell, researchers of the M4I Division of Imaging Mass Spectrometry investigate interfaces of crude oils using mass spectrometry. Crude oil is composed of the organic remaining of ancient organisms that lived hundreds of millions of years ago. Due to large variations in biological origin, in combination with prolonged exposure to high temperature and pressure in subterranean reservoirs crude oils form the most complex molecular mixtures that are found on Earth.
When a crude oil comes in contact with water or a rock surface some of the more polar molecules in crude accumulate on the boundary or interface layer between the oil and the water or between the oil and the mineral surface. These interactions make it difficult to extract oil from small pores in reservoir rocks, and often more than 60% of the oil remains stuck in a reservoir. Presently Shell does field experiments to extract more oil from reservoirs by using surfactants.
We study with imaging mass spectrometry which molecules from the crude oil tend to accumulate on the interface layer when oil comes in contact with water or with rock surfaces. The following research questions are taken into account:
- What type of oil molecules are at the interfaces?
- What is the thickness of interface layers?
- Is advantageous to prevent rock from being oil wet?
- Is there preferential association of oil to certain areas on rock?
- How are surfactant molecules distributed over the oil-water and oil-rock interfaces?
New sample preparation techniques
The development of new sample preparation techniques is the key element of this project. The molecular compositions of interface layers are studied among other things by secondary ion mass spectrometry (SIMS), laser desorption ionization (LDI) and desorption electrospray ionization (DESI) mass spectrometry.
Brainpath – Molecular Imaging
of Brain Pathophysiology
The Brainpath project carried out by the Marie Curie IAPP action brings together expertise on
in-vivo and molecular brain imaging. The project thrives towards building upon current developments in molecular imaging and creating an academic-industrial training and mobility framework to better understand brain disease and develop new preclinical strategies for early disease prognosis.
Within the consortium we, as one of the world leaders in high resolution mass spectrometry imaging, apply a multimodal mass spectrometry imaging (MSI) approach to identify new molecular biomarkers in early onset Alzheimer’s disease (AD), ischemia after induced stroke and brain plasticity during ontogeny of song birds. MSI has consistently demonstrated its unique capability of directly localizing a variety of biomolecules simultaneously from the tissue surface. By localizing specific neurotransmitters, lipids and peptides we can use these molecular signatures as important visualisation tools to study different stages of neurodegenerative disease progression and brain development. In collaboration with the University of Antwerp in Belgium, Leiden University Medical Centre in the Netherlands and Max-Planck Institute in Germany we are developing new strategies for structural investigation of biomolecules directly from their native cellular or tissue environment and correlating them with clinically established in-vivo positron emission tomography (PET) and magnetic resonance imaging (MRI).
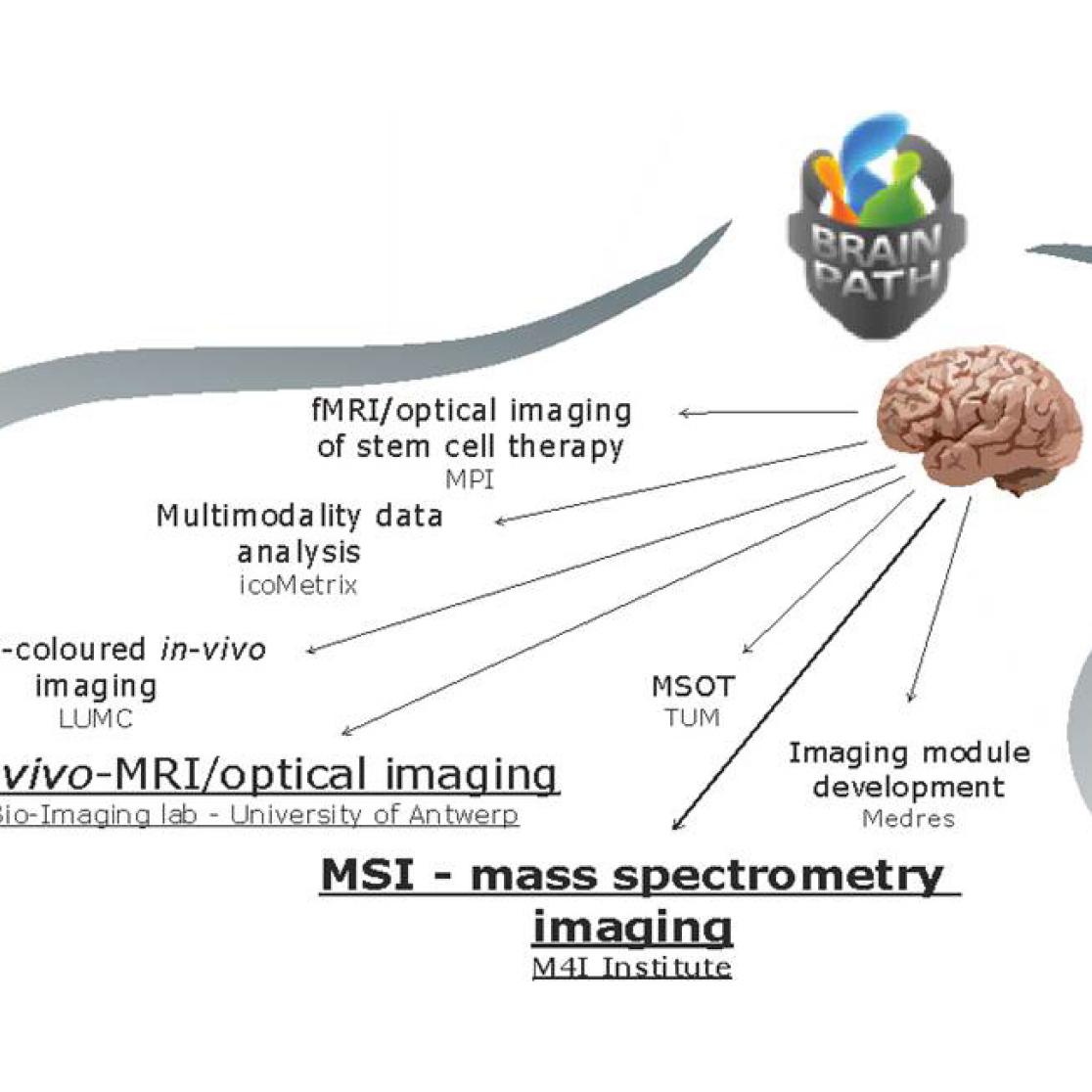
In-vivo imaging
With the concerted application of different high resolution MSI modalities in combination with in-vivo imaging we can provide a global insight into the different molecular structures involved in disease signalling, inflammation and tissue regeneration.
Visit our website Brainpath
Imaging hypoxia and tumor heterogeneity
Partner: Astra-Zeneca
In cancer research new drugs and combinations are tested vigorously in a preclinical environment in order to model the safety and effectiveness of new treatments. If the drug is effective in animal models, then these treatments are taken forward into clinical trials involving human patients.
To generate the preclinical model, usually modified cells from a human cancer are injected into a mouse and grown into a model tumour called a ‘xenograft’. Drugs are administered at carefully controlled doses and quantitative measurements taken of the tumour response. These measurements assume a uniform reaction across the mass of the xenograft. However both in preclinical models and clinical tumours the interior can be highly heterogeneous in both physiological and molecular properties; this is known as intra-tumour heterogeneity. This heterogeneity can drive very different reactions to the same drug within a single tumour. The loss of this detail in preclinical modelling may reduce the translatability of preclinical results into the clinical environment meaning that the effectiveness of new drugs can be misrepresented.
In this project we work with AstraZeneca to investigate molecular profiles that can be used to delineate regions of the intra-tumour heterogeneity. We focus on regions of low oxygen ('hypoxia’) due to leaky blood flow, and dead tissue that forms as a result of chronic hypoxia (‘necrosis’). Both of these factors have been shown to influence the behaviour and response of tumours to drugs in human disease. We use a combination of mass spectrometry imaging, optical imaging, and immunostaining in adjacent tissue sections to extract molecules that distinguish hypoxia and necrosis from viable tissue. For this we have developed a new analysis pipeline that can now be applied to other microenvironmental factors such as blood flow, or immune cell infiltrate. Hypoxia and necrosis - associated profiles are now being investigated in different cancer types to see if they can be consistently used to indicate these microenvironments.
Clinical testing?
The final goal is to integrate these profiles into the preclinical workflow to improve the detail included and the accuracy of the measurements taken to indicate whether a drug should or shouldn’t be taken forwards into clinical testing.
TargetCare: New approaches to joint diseases
Mobility is very important for human well-being, but is seriously impaired by osteoarthritis (OA) and intervertebral disc (IVD) degeneration in many people worldwide. This is mainly due to the degeneration of the cartilaginous tissue of joints.
The TargetCaRe project we are involved in, is part of Horizon 2020 Marie Sklodowska Curie Actions and involves 15 young scientists from 14 partner institutions from 5 different countries all over Europe, aiming to develop treatments for joint diseases by combining advanced drug delivery carriers with dedicated targeting tools and state of the art imaging techniques such as mass spectrometry imaging. The ultimate goal is to prevent further degradation of a damaged OA joint or IVD, as well as to activate the body’s own regenerative capacity to heal damaged and degenerated tissues.
In particular, we will be applying mass spectrometry imaging combined with in vivo imaging to provide proof of principle of the efficacy of new drug delivery systems and to identify cartilage degenerative and regenerative biomarkers. More precisely, TOF-SIMS will be used to study the distribution of the nanocarriers due to the high spatial resolution procured by this technique and MALDI-MSI to analyze the metabolite, protein and lipid regulation after compound administration and treatment with nanoparticles.
Visit our website TargetCare
PRISAR: Innovation in intra-operative diagnostics
PRISAR (pre-clinical intraoperative image guided surgery and postoperative radiotherapy of tumors) is supported by and carried out within the H2020 program RISE funded by the European Commission. The aim of the project is to promote skills development, transfer of knowledge and networking through cooperative development of intraoperative image guided surgery and postoperative radiotherapy of pancreatic tumors by hospital, academic and industrial groups throughout The Netherlands, United Kingdom, France and Germany. The scientific work packages aim to: 1) Develop a near-infrared labelled contrast reagent and tag with optimized affinity and selectivity for newly developed blood vessels; 2) Enhance visual detection of tumor margins by optimizing a Multi-Spectral Optoacoustic Tomography (MSOT) prototype, to detect tissue structures by sound and light) to an unprecedented tissue depth during small animal surgery; 3) Validate the fluorescent construct/antibody for integrin aub3 target combination across the subcellular, cellular, endoscopic and macroscopic levels; and 4) Capture locally applied radio-therapeutics after intraoperative image-guided surgery to demonstrate improved survival rates by improved radionuclide delivery to tumor without increase dose exposure to normal tissue. Additionally, cross-training schedules between 13 groups, transfer of knowledge and oversight of progress to meet deliverables of each work package is managed by Alan Chan of Percuros. Complete information on the PRISAR project H2020-MSCA-RISE grant number 644373-PRISAR.
Termed, molecular pathology, the information contributed to WP2 by M4I-IMS will assist in the ultimate goal of improving pancreatic tumor resection success for patients by providing complementary molecular information about the tumor/non-tumor interface from tissue obtained using image guided surgical techniques being developed in WP1 and WP2. M4I-IMS shares its results and findings not only with the PRISAR consortium, but also with members of the scientific community and general public. Knowledge sharing meetings, contributions to scientific conferences, hosting open days, hosting symposiums and cross trainings are a few examples of how M4I-IMS participates and leads in the skills development, transfer of knowledge and networking within and outside of the PRISAR consortium.
M4I-IMS at Maatricht Univerversity is involved in work package 2 (WP2) in conjunction with Ithera (pre-clinical MSOT) and Mauna Kea Technologies (Cellvizio) with the aim to investigate the tumor/non-tumor interface using mass spectrometry imaging techniques to identify minimum tumor margin distance based on molecular profiles.
Visit our website PRISAR
Imaging Mass Spectrometry in 3D digital pathology
Partners: ITEA, Philips, AMC, PS-MedTech
A strong growth forecast in the digital pathology market for the next five years combined with a decreasing number of qualified pathologists will lead to a tremendous increase in workload in the pathology departments of clinical and pharmaceutical organisations. On top of this there is an urgent need for higher quality diagnostic information enabling more effective and efficient treatments. The ITEA 3DPathology consortium will address these needs by creating a fast, digital, quantitative, spectroscopic and multimodal 3D pathology analysis system.
Imaging Mass Spectrometry will be an integrated part in this analysis system as it is expected to play an important role in future clinical routines for diagnostic and prognostic decision making.
Data analysis
As analytical partner in this ITEA project, M4I is leading the data acquisition work package and will provide multiple 3D imaging mass spectrometry datasets of clinical tissues and participate in the work package for data analysis.
Intra-operative diagnostics for
oncological procedures
Molecular information based surgery - Toward intraoperative MS-based
diagnostics for removal of hepato-pancreato-biliary (HPB) and breast tumors.
Partners: AZM-General Surgery & AZM-Department of Otorhinolaryngology |
Head and Neck Oncology
With more than 3 million new cases and 1.7 million deaths each year, cancer is one of the major health concerns in the modern European society, with continuously increasing incidence. Among the most prevalent forms are hepato-pancreato-biliary (HPB) cancers affecting the liver, pancreas and bile ducts; colorectal cancer (CRC); and breast cancer - the leading type of cancer in women. In most cases, surgery is the only curative treatment possibility, on the condition that the tumor has been diagnosed early enough. The patient, who is facing a life-changing diagnosis, has to undergo an invasive procedure with relatively high risk of recurrence, implying further surgical procedures with a low rate of long-term survival.
Improving the efficiency of surgery is bound to fundamentally improve the prognosis and quality of life for patients with the challenging diagnosis of cancer. The key hypothesis here is that accurate determination of the resection margin between tumor and non-tumor/healthy tissues and thus an improved rate of R0 resections in tumor surgery would reduce the recurrence rates in patients by lowering the risk of residual tumor tissue that remains during the intraoperative process. This would minimize removal and/or damage of healthy tissues as an undesirable effect for the patient and thus would potentially improve patient outcomes and survival rates. In collaboration with the Departments of Surgery and Pathology at MUMC+ we aim to deliver a novel molecular-guided platform for improving ex vivo cancer diagnostics and real-time diagnostics approaches during surgery. The first step will rely in building comprehensive molecular and histopathological classifiers (i.e. e-biobank) for HPB, CRC and breast cancers; which will potentially reveal new candidates for targeted therapies. The second step will be the incorporation of molecular-guided surgery tools for intraoperative MS-based diagnostics.

Identification of cancerous tissue
This innovative approach will present a more precise and accurate identification of cancerous tissue during surgical resection procedures and thus potentially improving the diagnosis and prognosis of cancer patients.
Metabolic profiling of bone during trauma surgery
Regenerative medicine in orthopedic trauma – toward the integration of bone and cartilage metabolic profiling in surgical setting. Partners: AZM-Orthopedics & MERLN
The incidence of fractures in the European population is increasing significantly due to the increasing number of elderly patients at risk. Of all fractures 20% needs to be treated surgically, involving an estimated 900,000 patients. Improvement in operative fracture exposure and fixation techniques has been substantial in recent decades, even for fixation of more difficult osteoporotic fractures in the elderly. It is estimated, however, that in up to 35% in high risk patients, fractures do not heal properly. In addition, at present, prediction of these post-traumatic complications – such as osteoarthritis –at fracture surgery is inaccurate, and not based on bone metabolism and the delayed presentation of complications further compromizes adequate treatment. There is therefore an urgent clinical need of new and innovative approaches to improve surgical procedure and patient’s management.

In close collaboration with the clinical research groups of the orthopedic and trauma surgery departments at azM and MERLN we investigate new MS based surgical tools to study bone and cartilage metabolism. This approach will provide us with an in situ and in real-time diagnostic technique and guide newer therapies aimed at regeneration of the bone and cartilage.
Molecular histology with imaging
MS for colorectal cancer studies
Partner: AZM-Pathology
Colorectal cancer is the third most common cancer worldwide with over a million new cases diagnosed every year. Due to the implementation of CRC screening programs, the number of early stage CRC cases is increasing steadily. However identifying which of these cases are especially high risk, or would benefit from specific treatments such as neoadjuvant therapy (chemotherapy before surgery), is extremely challenging. The tumour node-metastasis staging (TNM) system, which rates the characteristics of the primary tumour, and invasion of nearby lymph nodes and distant metastasis, is the current gold standard, but performs sub-optimally in these early stage cases.
Many other clinico-pathological and molecular characteristics are therefore under investigation for their prognostic and predictive value. These include detailed investigations into tumour characteristics, surrounding lymph nodes, and metastasis, as well as genetic factors such as microsatellite instabilities, DNA methylation, and mutation and expression profiles. Although several show promise, none have yet been implemented clinically. Either the prognostic value is too low, or a standardised definition of the feature is unavailable, or, most commonly, the markers lack appropriate validation and testing in large, non-biased cohorts with proper statistical testing to prove their additional prognostic value in comparison to the current TNM system.
In this collaboration we apply mass spectrometry imaging to analysing the molecular composition of hundreds of CRC patient samples, in combination with full clinical data, staining, genomics, and immunohistochemistry. The molecular profiles will be investigated for potential CRC tumour subtyping.
Analysing the molecular composition
Evidence suggests that several types of CRC cancers exist, each with distinct prognostic characteristics, however a reliable system for distinguishing these has not yet been discovered. Furthermore the full complex data will be carefully analysed to develop a multivariable prediction model of prognosis for various clinical factors including 5 year survival and CRC staging. Further cohorts are available to validate the model, assisting its clinical acceptance, and giving this project a unique opportunity to support individualised treatment and treat high risk CRC cases at an early stage.
Molecular imaging of
osteoarthritic cartilage and regeneration
Partners: AZM-Orthopedics, University Twente, DTL, ZonMW
Osteoarthritis (OA) is the most prevalent form of arthritis. OA affects 80% of the population over the age of 65. It is a complex pathology because diverse factors interact causing the process of deterioration of the cartilage. The current view is that OA is a group of diseases that can be differentiated based on the risk factors and on the pathophysiological mechanisms underlying the joint damage. However, current trials with potential therapies do not distinguish between different subtypes of OA.
Application of MSI in the field of drug delivery also provides insight in the relation between tissue distribution, activity in target tissue and possible local side effects in other tissues, e.g. osteoporotic bone changes, small molecule and tissue characteristics determining penetrance. Local controlled delivery of drugs circumvents side effects of systemic administration while reducing dosing height and frequency. The effectivity of drug delivery systems depends on their release kinetics and dosage. Typically, in the preclinical stage of development, this is measured by determining serum levels or, if available synovial fluid. However, serum levels do not reflect tissue content, as penetration is highly dependent on tissue and drug characteristics. In collaboration with UMCU we aim to reveal the distribution of two anti-inflammatory drugs released from locally injected microspheres in joint tissues, such as cartilage, tendon and bone by MSI. This approach will allow for further fine-tuning of the controlled release-drug combinations and understanding of the principles of controlled release key in the development of efficient drugs. With the support of DTL-associated Technology Hotels and Life Sciences Health funding (ZonMw) we expect to obtain a head start on this novel crossroads between medical biology, drug delivery and molecular imaging.
Mass spectrometry imaging (MSI) has become a powerful method for tissue-based disease classification
The power of MSI resides in the ability to detect lipids, proteins, drugs, metabolites and other compounds directly from tissue whilst preserving the information about their spatial localization. Preliminary work of our group has classified OA and healthy groups based on the peptidomic and lipidomic profiles of human cartilage and synovium.
In collaboration with the Orthopedic Surgery department at MUMC+ and the division of Rheumatology at azM we aim to identify different OA phenotypes by MSI and use this novel high throughput strategy to reveal new markers for a personalized medicine.
Intra-tumor heterogeneity
and personalized medicine
Partners: Helmholtz Zentrum Muenchen, Munich, Germany, Fondazione della Scienza ONLUS, Pisa, Italy, West German Cancer Center, Essen, Germany, Klinikum rechts der Isar, Munich, Germany, Institute of Genetics and Molecular and Cellular Biology, Strasbourg, France.
In many tumors, including esophageal and esophagogastric junction adenocarcinomas (EAC), intra-tumor molecular heterogeneity is closely linked to tumor characteristics that are responsible for a poor prognosis of the patients such as therapy resistance, early metastasis and tumor recurrence. Hence there is a strong need to identify clinically-relevant tumor subpopulations.
In this study, we will use Imaging Mass Spectrometry to investigate intra-tumor molecular heterogeneity in EAC with the aim to identify robust molecular classifiers which help to direct therapy based on an analysis of sequential tumor material from the MEMORI trial (Metabolic and Molecular Response Evaluation for Therapy Individualization in esophageal and esophagogastric junction adenocarcinomas, EUDRA-CT Number 2014-000860-16). In this clinical trial, 14 days before and after induction of neoadjuvant chemotherapy tumor response is evaluated using PET and sequential tumor biopsies obtained. This setting gives the unique opportunity to study the evolution of intra-tumor heterogeneity of EAC in the context of therapy response through a molecular examination of the tissue specimens before, under and after neoadjuvant therapy.
Multiplex molecular information
Imaging Mass Spectrometry has the unique capability to decipher intra-tumor heterogeneity as it allows reading out multiplex molecular information of tissues in a spatially-resolved way without any label.
Unraveling the complexity of troponins in human heart
by imaging mass spectrometry
Partner: azM Clinical Diagnostic Laboratory
Troponin is a unique and heart specific protein (troponin T or I) and is released into the bloodstream after an infarction. Using the recently introduced high sensitive troponin immunoassays it is also possible to detect troponin in healthy individuals. Increased troponin concentrations are found in diseases like heart failure, myocarditis, renal failure and also endurance exercise. This complicates the anamneses of the typical and atypical chest discomfort to a cardiac cause.
Here, we target the investigation of the release of cardiac troponin by cardiomyocytes, using clinical diagnostic tests and imaging mass spectrometry. Focusing on understanding the local molecular changes in myocardial dysfunction on one hand and unraveling metabolic patterns and pathways of troponin release on the other hand. Native mass spectrometry can be used as well to investigate the conformation of the troponin complex with and without the effect of Ca2+ and/or implicated enzymes. All together this can give insight in the time dependent degradation pattern of troponin, a possible correlation with the stability of atherosclerotic plaques or the diagnostic distinction between the acute and chronic elevation of troponin concentrations.
Unraveling metabolic patterns
Focusing on understanding the local molecular changes in myocardial dysfunction on one hand and unraveling metabolic patterns and pathways of troponin release on the other hand.
Forensic hair testing
Hair testing is a powerful tool routinely used for the detection of drugs of abuse in toxicology and forensic applications. The analysis of hair is highly advantageous as it can provide prolonged detectability versus that in biological fluids and chronological information about drug intake based on the average growth of hair. However, current methodology routinely involves complex and time-consuming sample preparation followed by gas or liquid chromatography coupled with mass spectrometry. This project has investigated the use of mass spectrometry imaging techniques to visualise the distribution of drugs of abuse (such as cocaine) in single human hair samples, these techniques offer the ability to provide more accurate and visual chronological information on drug intake (hours to days) over current techniques which only provides values on a monthly basis.

These techniques have also been used to investigate the effects of current forensic decontamination protocols and cosmetic treatment on the analysis of drug user hair samples.
Quantitative molecular imaging
in pharmaceutical sciences
In the pharmaceutical industry Mass Spectrometry (MS) is an integral analytical tool used throughout novel drug research, e.g. in discovery, DMPK, in pre-clinical and clinical toxicology, impurity profiling, stability testing, efficacy testing etc. The use of Imaging MS though us still relative new and visualizing spatial distribution of an active drug and associated metabolites or complete affected metabolic pathway offers still a lot of challenges. On the other hand spatial information of new (and existing) drugs and associated metabolites in tissues will improve mechanistic understanding and clinical consequences of pre-clinical safety concerns and will direct better decision making in further drug development. Besides, local spatial and quantitative information will allow PK/PD assessment at the tissue level as well as target/pathway engagement. In combination with MS based (metabol)omics it represents a highly promising approach in mechanism based safety evaluation as well as (novel) target identification/validation and will prove to be extremely valuable across all therapeutic areas.
Severe challenges
As mentioned though there are still severe challenges, like e.g. attainable spatial resolution of low to sub um level, reliable quantitative information and improved sensitivity of IMS. Therefore, we have shaped three separate projects to address these challenges and at the same time apply the developed technology in relevant projects and real-life examples.
Project Eurlipids - Interreg EMR
This Interreg EMR funded project aims to create an internally well recognized knowledge center in the field of Lipidomics, by smartly and successfully crossing the borders in technical and biomedical science AND countries. The consortium will develop educational master courses at different universities, along with the creation of a Lipidomics screening infrastructure. The focus areas in the project are on advances in mass spectrometry-based imaging and profiling technologies and their application for neuro-degeneration (Alzheimer and Multiple Sclerosis) diseases, cancer and Cardio-Renal disorders.
Our overall goal is the generate breakthroughs in the causality between lipid profiles and the on-set and progression of diseases.
We target the development of new ionisation technologies, introduction of ion mobility spectrometry, new statistical data processing and bioinformatics approaches and even so important improve sample selection and sample pre-treatment procedures for the quantification and identification of large families of lipids. Here both blood/plasma and tissue samples from animal models and patient cohorts are investigated.
As an economic output, the project will enable the commercialization of the new technologies and methodologies developed; making the lipidomics platform available for researchers outside of the consortium.
Likewise, reaching out to patient organizations and jointly defining new challenges in health-care and lipid research is essential in achieving a sustainable and reliable partner in health-care and life-sciences innovations.
Our overall goal is the generate breakthroughs in the causality between lipid profiles and the on-set and progression of diseases.
More information: Eurlipids or Interreg EMR
