- Analysis of human molecular Big Data (epigenomics, transcriptomics) in the context of cardiovascular disease, using existing computational tools.
Research objectives
The key objective of the M4I Division of Nanoscopy is to gain greater insight into the 3D form of cell proteins, thus paving the way for the development of more effective treatments for diseases such as cancer and tuberculosis. A better understanding of how protein complexes manage healthy, but also diseased, cells will allow drugs and vaccines to target problems more effectively.
For instance, professor Peter Peters and his team are working to improve the vaccination presently used against tuberculosis. This research is based on Peters’s finding in 2007 of how the bacteria that cause tuberculosis behave in within cells. Each day, several thousand people die of tuberculosis, especially in developing countries. The M4I Division of Nanoscopy also aims to develop greater insight into the workings of the immune system, which may in the future lead to an immune response against cancer cells in the human body.
Ongoing projects
- Project 1: T7SS
- Project 2: Cryo-electron tomography
- Project 3: Nanofluidic chamber
- Project 4: Linezolid effects
- Project 5: Technology improvement
- Project 6: IN-SENSE
- Project 7: Model tuberculosis
- Project 8: Non-animal technology development
Aeras and NLF: Type VII secretion system in Mycobacterium tuberculosis
Project 1 T7SS
In 2007 our team published an article in Cell showing that after prolonged infection in macrophages and dendritic cells, M. tuberculosis translocated from phago-lysosomes to the cytosol and killed the host cell a few days later, while the BCG vaccine strain failed to translocate. We found that this process was dependent on a gene in the extended RD1 region (ext-RD1). The translocation to the cytosol was unexpected, as it contradicted the prevailing dogma. We then focused on BCG with a knock-in of the entire ext-RD1, as this region is now known to encode the ESX-1 secretion system, which was recently identified by one of our former team member as a novel type VII secretion system (T7SS) (Abdallah AM et al., Nat Rev Microbiol. 2007; see figure below). We found that these bacteria translocate to the cytosol of the host cell 7 days after infection. We concluded that the ESX-1 system (in a BCG background) is sufficient for translocation (Abdallah AM et al., J of Immunol. 2011 and Houben et al., Cell Microbiol. 2012). Therefore we believe that the T7SS is the key to virulence in tuberculosis. Using cryo-EM, we therefore investigated the structure of the mycobacterial capsular layer and showed that high levels of the ESX-1-secreted proteins were present in the capsule (Sani et al. 2010).
We are now using cryo-EM single-particle analysis (SPA), and over the next years will investigate recombinant purified proteins of individual gene products from the T7SS. In addition, we are purifying the entire intact T7SS structure (or mutants thereof) using biochemical methods for 3D reconstruction. Our ultimate aim is to image mycobacteria in vitreous sections in human host cells while translocating. The cryo-SPA, X-ray, and NMR data of the T7SS can then be docked into the images from vitreous sections to construct a macromolecular map of the tubercle bacillus within the host cell. The broader objective is to gain insight into the structure and function of the mechanism for type VII-mediated translocation. This should lay the groundwork for the development of novel antibiotics and better vaccines.
Plunge freezing will be used to obtain a thin vitrified film containing isolated mycobacteria whose outer cell wall has been removed. This will then be subjected to cryo-EM imaging. Purified protein complexes composed of RD1-gene products will be obtained from Stewart Cole in Switzerland, Ravi Ravishankar in India and other scientists worldwide, and subjected to SPA as well as tomographic imaging. This process will give rise to 3D images that can be used as models for host-pathogen interaction. The approaches will be developed with Mycobacterium BCG: RD1 in human cells (adapted to biosafety level 2) as a model for host-pathogen interaction, leading to insights that can be used in the development of new tuberculosis vaccines and drugs. An ERC advanced grant draft for this project is currently being prepared.
STW: Cryo-ET with cryo-SR-LM: new tools for imaging nanomachines in cells
Project 2 Cryo-electron tomography
Cryo-EM currently provides the ultimate resolution in single-particle analysis (SPA) (<0.5 nm) and in cryo-ET (<2 nm). However, the high information content and low contrast of cryo-EM images often makes it impossible to locate the subcellular structure of interest for cryo-ET. One possible solution may be state-of-the-art correlative light/electron microscopy (CLEM) studies. In these studies, interesting cellular features are identified by imaging GFP-tagged proteins (available for almost all gene products of the genome) using live-cell microscopy, followed by rapid immobilization and cryo-ET. Put simply, cryo-ET provides the extreme resolution needed to construct the detailed 3D image, whereas the proteins are identified by their genetically encoded fluorescent labels. Currently, the limited resolution (~250 nm) of conventional fluorescence microscopy makes it impossible to unequivocally localize the fluorescent labels to the nanomachines.
To develop this solution, we intend to modify the FEI dedicated cryo-LM holder capable of maintaining the frozen-hydrated EM grid in its vitrified state (recently developed at the Max Planck Institute in Martinsried) for use on the super resolution LM such that it can work under cryogenic conditions. Using this setup, we will then apply PALM principles to obtain optical confocal and later super resolution (SR) cryo-images that identify individual proteins in nanomachines at particular subcellular sites. The preparation will subsequently be transferred to the cryo-EM under cryogenic conditions for ET imaging at sub-nanometer resolution. We will screen the samples with a cryo-TEM (200kV) to select those suitable for cryo-ET at NeCEN. Finally, we will deliver combined image modalities with upwards of 20nm correlation accuracy to identify individual macromolecular machines, which should provide a glimpse into the molecular makeup of the cellular complexes at unprecedented resolution levels.
Successful completion of this task depends on development of several new tools:
An existing cryo-LM holder will be adapted to fit the SR microscope. This is a challenging task because the potential thermal drift on the stage, caused by liquid nitrogen in the cryo-LM holder, must be kept to an absolute minimum. Further, as immersion oil cannot be used at these temperatures, we will employ a 100 x, 0.9 NA dry objective (Fig. 1).
We will further develop the MAPS navigation tool, which allows for correlation of the SR images with the cryo-EM images with a relocation precision greater than 50 nm. This requires updating of the existing software to high standards and the introduction of fiducial markers.
This project will benefit from a parallel project to develop novel cryo-ultramicrotomy. In particular, we will collaborate with Diatome to adapt their (room-temperature) oscillating diamond knife to obtain cryo-sections with almost no compression in the preparation.
The project will also benefit from another parallel side project with Maastricht Instruments. Briefly, because intact cells are too thick to be imaged by means of EM without sectioning, we will develop an alternative method called the “double grid approach”. In this device, the cells spread voluntarily to ~200 nm thickness, and can be vitrified with our Vitrobot for ET imaging in a close-to-native state. Maastricht Instruments will integrate an objective lens inside our Vitrobot to observe the migrating cells on a golden EM support grid and manufacture “double film” EM grids. The imaging and screening of these thick preparations by way of cryo-EM requires a 200 kV.
The developments described here will lead to a better understanding of the positions and states of nanomachines in normal and diseased cells. We will base validation studies of the new instrument on characterization of the new type VII secretion system (see project 1) involved in this translocation. The advances from these studies will greatly benefit not only cryo-ET, but also the SR imaging community. For example, electron microscopists have long known that chemical fixation methods introduce structural artifacts at tens of nm, whereas SR microscopy in STED and PALM mode is routinely performed on formaldehyde-fixed material. This limits the interpretation of high-resolution SR images from a biological perspective. Cryo-SR LM will image proteins in a much closer-to-native preparation. Importantly, the rate of fluorophore bleaching at cryogenic temperatures will be significantly reduced, which means more photons will emit from each fluorophore, resulting in increased resolution. In theory, this effect should far outweigh the lower light-collecting properties of the dry objective.
To achieve the goals laid out in the STW proposal, industry practitioners must develop new tools in collaboration with academic partners. The interest among companies in realizing this aim is evident from their generous offers to contribute. The project will facilitate the development of commercial products that will be in high demand among cryo CLEM labs (academic and industrial) worldwide; namely, a cryogenic chamber, the integration of this chamber with a state-of-the-art SR microscope, and tools to facilitate rapid relocalization of the preparation from the SR setup to the EM.
NanoNext: Nanofluidic chamber for cryo-ET
Project 3 Nanofluidic chamber
Cryo-ET of vitreous sections is an emerging technique that produces high-resolution (2–4 nm) structures of macromolecular complexes in frozen-hydrated state within their native environment. High-pressure frozen cells are sectioned into long ribbons 50nm thick, and viewed using cryo-EM. This approach is technically demanding, and it is difficult to ascertain whether the macromolecular complexes are disrupted by the freezing and sectioning processes. While it has been shown that whole bacterial cells can be visualized without sectioning (Kühner et al. Science 2009), the electron scattering prevents high-resolution imaging for organic samples thicker than 200 nm, which renders mammalian cells (at several microns) too thick. As an alternative to vitreous cryo-sectioning, we recently showed by way of live-cell light microscopy that migrating white blood cells (DCs, T-cells, etc.) can naturally migrate into a 1000 nm thick “nanochamber”. Using nano technology, we now intend to develop a method to vitrify the cells in this state. They are then sufficiently thin to be subjected to a cryo-FIB/SEM procedure making them 200nm, and thus useful for cryo-ET studies of macromolecular complexes within native mammalian cells without the need for cryo-sectioning.
The M4I Division of Nanoscopy will develop this chamber with silicon-based microelectromechanical systems (MEMS) technology in collaboration with colleagues at TU Delft. Two main challenges need to be addressed. First, a chamber must be designed that is even more transparent than the cell, yet still mechanically robust. This chamber should fit seamlessly with the existing high-tech equipment: cryo-EMs, cryo-fluorescent microscopes, and cell preparation tools. Second, high throughput methods must be developed to use the chamber efficiently and obtain resolution within the nanometer range. The envisaged device will allow us to study the behavior of intact mammalian cells and their responses to drug-induced changes at the molecular level. The project is ‘high risk’, but could open up new avenues for nanoscale research on intact mammalian cells. If successful, we intend to investigate human dendritic cells infected with M. marinum as well as the macromolecular interaction of T7SS with the phagolysosomal membrane.
EU FP7 Notox: Effects of linezolid on ribosomes of M. tuberculosis: a structure-function study
Project 4 Linezolid effects
At present, drug-induced changes of macromolecular targets cannot be observed within the native cellular environment. Ideally, a technique must be developed that allows for the observation of high-resolution macromolecular structure changes within this environment. One promising antibiotic for the treatment of multi-drug resistant (MDR) tuberculosis is linezolid.
Linezolid acts by inhibiting bacterial protein synthesis. The available X-ray crystal structures of linezolid in complex with 50S ribosome of Haloarcula marismortui (an archaeon) suggest that linezolid may inhibit the formation of the ribosomal initiation complex (Joseph et al., J Med Chem. 2008). As yet, however, the mechanism of action of linezolid on M. tuberculosis has not been definitively identified. This project focuses on the effect of linezolid on ribosomes isolated from mycobacteria, with a view to answering the following questions:
- Does linezolid prevent the formation of functional mycobacterial ribosome initiating complexes in vitro?
- Does linezolid have the same mechanism of action in vivo (within the context of the entire bacterial cell)?
We will evaluate the quality of the ribosome preparation by means of cryo-EM. Using the same technique, we will also study the effect of linezolid on the formation of mycobacterial 70S ribosomes and determine the optimal concentration for inhibiting complex formation. Subsequently, we will investigate the effect of linezolid on in vitro purified mycobacterial ribosomes and on M. tuberculosis cells to determine the minimal inhibitory concentration of antibiotic that has a bactericidal effect on our strains. To this end, we will use fluorescent microscopy to observe bacterial cell death with Live/DEAD BAC/Light assay. Finally, we will prepare M. tuberculosis treated with linezolid for cryo-EM of vitreous sections using high-pressure freezing (HPF) and vitreous cryo-sectioning techniques (Pierson et al. J Structural Biology 2011) or cryo-FIB/SEM lamellae to identify the mechanism of action of drugs on ribosomes inside entire bacterial cells.
This project is intended as a prototype for structural characterization of mycobacterial ribosome. It will provide the research team with scripts and protocols for SP-cryo EM designed to tackle specific issues that may arise during ribosome reconstruction.
New technology improvement for the Titan Krios cryo-EM: phase plate
Project 5 Technology improvement
If a specimen merely changes the phase of the electron wave (i.e. the specimen is a phase object), provided the electron microscope is at zero focus and there are no aberrations, there will be nothing to observe. That information was obtained from biological specimens in the past was partly due to the poor optical performance of the TEMs, and partly due to the use of a relatively large defocus. However, a significant amount of information is then lost. Contemporary electron microscopes can be equipped with an aberration corrector, and at zero focus on a phase object virtually nothing will be observed. Yet switching to off-focus also results in a loss of information. Thus, a phase plate – similar to the Zernike phase plate in light microscopy – would clearly be desirable. This phase plate should effect a phase shift of pi/2 on the central beam compared to the diffracted beams, and should not block the relevant electrons. In biology, all information beyond 0.01 Å – apart from the phase-shifted central beam – should be passed up to 1 Å through the phase plate. The phase plate must be located in the rear focal plane of the TEM. It will allow all phase information to be translated into amplitude information, which will vastly improve the imaging of biological specimens.
Several research groups have experimented with phase plates. Two types of phase plates can be identified: a) a thin sheet of C, for instance, can be placed such that all diffracted beams have a phase shift of pi/2; or b) the central beam is led through a small tube that effects a phase shift on the central beam. The second approach is the more appropriate. However, although such phase plates have been used successfully for over 5 years, no commercial product is yet available. This can be attributed to the difficulties involved in developing them. Not only must they be extremely small (with ~50 nm precision); the issues of contamination and charging also pose problems. In collaboration with Henny Zandbergen and his team at the Kavli Nanoscience Institute (TU Delft), we have performed tests showing that we can make apertures with the required degree of precision. As we also have solution to the contamination and charging problems, we aim to develop a reliable phase plate of the appropriate dimensions.
FP7 EU: IN-SENSE; Deciphering inter cellular signalling in schizophrenia
Project 6 IN-SENSE
Chronic mental illnesses (CMI) like schizophrenia or the recurrent affective disorders are among the most debilitating (prevalence 1-10%) and costly diseases in the Western world. Research on brain diseases is a focus in FP7. In fact, direct healthcare costs of the two disease categories that IN-SENS is directly addressing, the psychotic and affective disorders cost four times as much as dementia and other neurodegenerative disorders. This is mainly due to the early onset of CMI in adolescence which leads to in most cases lifelong impairments and occupational disability. Yet, even though CMI have dramatic impact on individual patients, their relatives, and society, our biological understanding and, accordingly, our options for novel strategies of efficient treatment or cure of CMI are stalling.
The last decade has brought breakthroughs in the genetics of CMI, like the discovery of the disrupted-in-schizophrenia 1 (DISC1) gene (by a member of this consortium), and others. The identification of these candidate genes now allows analysis of the INter- and intracellular Signalling INSchizophrenia (hence the project acronym IN-SENS). A European perspective is necessary since in most EU countries the infrastructure for the necessary interdisciplinary research is not met on national levels. In Maastricht we will be doing cryo-immunogold EM to localize DISC1 in the healthy and diseased brain.
Zon-MW: Human in vitro tissue and single cell approaches to model tuberculosis
Project 7 Model tuberculosis
Start date 2014
An urgent task of today’s medical research is to find new ways to treat infections caused by Mycobacterium tuberculosis. There are numerous reports on multidrug-resistant strains of this pathogen, which causes tuberculosis (TB). Most experimental studies of TB rely on animal experiments and there are no existing alternative models such as in vitro tissue models, with the exception of infected single cell cultures. Replacing existing animal models with human in vitro systems is also important because of the questioned validity of these models for TB. The aim of the presented project is to further develop two novel human in vitro models for TB, validate them against tissues from TB patients and to use them instead of animals for testing anti-TB combination regimens based on existing drugs. The models are based on human primary cells and cell lines and the investigation of these models as substitutes for animal models for preclinical drug testing.
The first model is based on macrophages obtained from human blood that are infected with virulent M. tuberculosis. Similar models have been described, but the unique feature of the presented model that the model displays is that under the right conditions, it can mimic clinical latent TB. Understanding latent TB is of importance, since the latent infection remaining in patients on TB treatment is very tolerant towards antibiotics and is causative of multidrug resistance. The so far confirmed results include absence of net growth of the bacteria inside the macrophages over two weeks, alteration of characteristics of the bacteria identical to observations in other studies, suppression of immune activation of the infected cells, tolerance towards antibiotics and most importantly, regrowth of the bacteria during immunosuppression, which is a well-known clinical problem and has so far only been modeled in animals. The second model is based on the establishment of co-cultures of human cell lines and primary immune cells on a filter with a collagen matrix. Human blood monocyte-derived macrophages infected with M. tuberculosis are introduced into the system. The tissues are exposed to air on the apical side, causing the epithelial cells to secrete mucus, further mimicking the microenvironment in the lung. This unique TB model will be characterized within the project. During the present project, the models will be compared with and validated against human TB-infected tissues on an ultrastructural level using advanced electron microscopy (Maastricht University). The proposed project is a translational academic project linking experimental research to clinical testing and including Dutch and Swedish research groups. The models will be used for screening combinations of approved drugs together with TB-antibiotics and for achieving proof-of-principle of adjuvant treatments for a more effective TB treatment. Together, the two models may prove to serve as good substitutes for animal models for TB. If the project is successful, the models can be further developed to mimic different kinds of bacterial and viral infections.
Non-animal technology development
Project 8 Non-animal technology development
Collaboration with Hans Clevers.
Project proposal in preparation
NC3Rs – National Centre for the Replacement, Refinement and Reduction of Animals in Research
Advancing the development and application of non-animal technologies - NEW COMPETITION FOR FEASIBILITY STUDY FUNDING
read more on NC3Rs
- Project 1: High throughout spectral imaging with pixelated detectors
- Project 2: Polyimage: Innovations in ambient imaging mass spectrometry
- Project 3: Imaging interfaces in geochemistry
- Project 4: Brainpath – Molecular Imaging of Brain Pathophysiology
- Project 5: Imaging hypoxia and tumor heterogeneity
- Project 6: TargetCare: New approaches to joint diseases
- Project 7: PRISAR: Innovation in intra-operative diagnostics
- Project 8: Imaging Mass Spectrometry in 3D digital pathology
- Project 9: Intra-operative diagnostics for oncological procedures
- Project 10: Metabolic profiling of bone during trauma surgery
- Project 11: Molecular histology with imaging MS for colorectal cancer studies
- Project 12: Molecular imaging of osteoarthritic cartilage and regeneration
- Project 13: Intra-tumor heterogeneity and personalized medicine
- Project 14: Unraveling the complexity of troponins in human heart by imaging mass spectrometry
- Project 15: Forensic hair testing
- Project 16: Quantitative molecular imaging in pharmaceutical sciences
High throughout spectral imaging
with pixelated detectors
This STW project aims to develop new direct imaging detector systems based on the Medipix/Timepix chip for applications in X-ray, ion and electron imaging. The focus areas in biomedical imaging are molecular histology with mass spectrometry and low-dose, energy dispersive X-ray imaging. We target the development of an ion imaging system that is suitable for the detection of macromolecular ions of either polarity, which can be post-accelerated up to 15kV. In this way, we create a bio-molecular imaging device for molecular histology for use with ion mobility and time-of-flight mass spectrometers. A complimentary imaging approach aims at energy dispersive X-ray imaging. A novel system will be developed with a new MPX chip based device that makes it possible to separate the incoming X-rays based on their energy and use eight thresholds to make seven windows in which one can count the incoming photons. Our overall goal is to improve quality and reduce the measuring time in ion and electron imaging and introduce energy information into X-ray imaging and thereby reduce the required dose.
Microscope mode mass spectrometry imaging using a pixelated detector
Bare chips of the Timepix type in combination with microchannel plates (MCPs) are implemented on a triple focusing time-of-flight (TRIFT) mass spectrometer as a detector for microscope mode mass spectrometry imaging of organic molecules in biological samples at high speed. (Figure 1) Applying a high potential on the Timepix/MCP detector, a wide mass range and an increased signal-to-noise ratio can be achieved compared with other microscope mode instruments. [1-6]
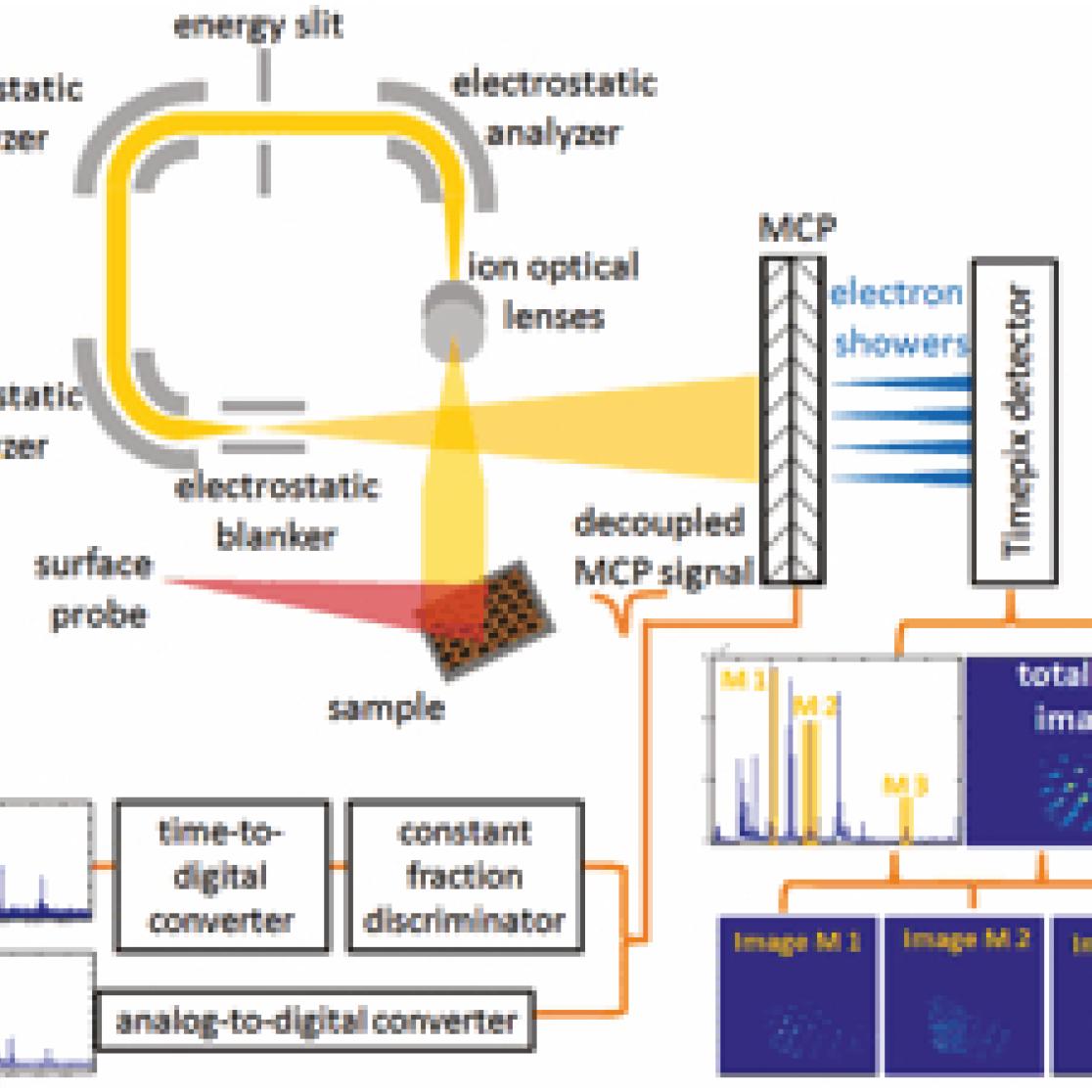
Figure 1 – Schematic overview of SIMS-trift ion microscope setup with implemented MCP/Timepix detector. Mass spectra are acquired without the need of mass selection. The spatial distribution of compounds on the sample surface are maintained during ionization and mass seperation and projected on the Timepix detector as such.
Polyimage: Innovations in ambient
imaging mass spectrometry
PolyImage - The development of a new chemical microscope for research of polymers and smart materials. Partners: Wageningen University, Maastricht University, DSM Resolve, Waters Chromatography, Omics2Image and RIKILT.
MSI is a technique in which molecules are locally extracted from a surface and ionized (electrically charged) after which their mass can be determined. The mass is then used to determine the molecular structure, which provides us with surface images of molecular distributions on top of the structural information provided by e.g. optical microscopes. This way the distribution of drug molecules and metabolites can be followed in tissue without labelling, or molecular surface changes to synthetic polymers can be monitored, for example.
Currently, MSI is not able to provide all molecular information from every type of sample: Many samples of interest, such as unfrozen tissue, unprocessed food or water containing polymers, are not compatible with high vacuum, which is needed for most current MSI methods as it provides much higher sensitivity. Liquid containing samples cannot be introduced into high-vacuum as the liquid would immediately evaporate. These samples are of high interest to both science and industry, for example to explain disease processes or improve food quality.
Molecules relevant for investigation, such as polymer additives or enzymes, are often present in relatively low abundancy, which makes the detection of these molecules much less likely. To be able to detect all molecules, the ionization method needs to be able to charge molecules along the full range of physical properties. For example, molecules with low proton affinities are not ionized by electrospray (attachment of a proton), which is the most efficient and common way of ionizing molecules at atmospheric pressure. In contrast, plasma ionization (detachment of an electron) is selective to molecules with low proton affinities.
Technical and chemical improvements
We aim to devise an instrument able to apply MSI on liquid containing samples with high sensitivity while also having the possibility to detect molecules with low proton affinities. Technical and chemical improvements in mass spectrometry imaging instruments must enable researchers to study the surface of (bio)polymers and smart materials more precisely. This new instrument will be more sensitive and applicable for measurement of chemicals on end products and intermediates, for trouble-shooting of products and production processes, for the development of new natural plastics, for the management of conversion of biomass and for the improvement of biosensors.
Imaging interfaces in geochemistry
Partner: Shell
In collaboration with Shell, researchers of the M4I Division of Imaging Mass Spectrometry investigate interfaces of crude oils using mass spectrometry. Crude oil is composed of the organic remaining of ancient organisms that lived hundreds of millions of years ago. Due to large variations in biological origin, in combination with prolonged exposure to high temperature and pressure in subterranean reservoirs crude oils form the most complex molecular mixtures that are found on Earth.
When a crude oil comes in contact with water or a rock surface some of the more polar molecules in crude accumulate on the boundary or interface layer between the oil and the water or between the oil and the mineral surface. These interactions make it difficult to extract oil from small pores in reservoir rocks, and often more than 60% of the oil remains stuck in a reservoir. Presently Shell does field experiments to extract more oil from reservoirs by using surfactants.
We study with imaging mass spectrometry which molecules from the crude oil tend to accumulate on the interface layer when oil comes in contact with water or with rock surfaces. The following research questions are taken into account:
- What type of oil molecules are at the interfaces?
- What is the thickness of interface layers?
- Is advantageous to prevent rock from being oil wet?
- Is there preferential association of oil to certain areas on rock?
- How are surfactant molecules distributed over the oil-water and oil-rock interfaces?
New sample preparation techniques
The development of new sample preparation techniques is the key element of this project. The molecular compositions of interface layers are studied among other things by secondary ion mass spectrometry (SIMS), laser desorption ionization (LDI) and desorption electrospray ionization (DESI) mass spectrometry.
Brainpath – Molecular Imaging
of Brain Pathophysiology
The Brainpath project carried out by the Marie Curie IAPP action brings together expertise on
in-vivo and molecular brain imaging. The project thrives towards building upon current developments in molecular imaging and creating an academic-industrial training and mobility framework to better understand brain disease and develop new preclinical strategies for early disease prognosis.
Within the consortium we, as one of the world leaders in high resolution mass spectrometry imaging, apply a multimodal mass spectrometry imaging (MSI) approach to identify new molecular biomarkers in early onset Alzheimer’s disease (AD), ischemia after induced stroke and brain plasticity during ontogeny of song birds. MSI has consistently demonstrated its unique capability of directly localizing a variety of biomolecules simultaneously from the tissue surface. By localizing specific neurotransmitters, lipids and peptides we can use these molecular signatures as important visualisation tools to study different stages of neurodegenerative disease progression and brain development. In collaboration with the University of Antwerp in Belgium, Leiden University Medical Centre in the Netherlands and Max-Planck Institute in Germany we are developing new strategies for structural investigation of biomolecules directly from their native cellular or tissue environment and correlating them with clinically established in-vivo positron emission tomography (PET) and magnetic resonance imaging (MRI).
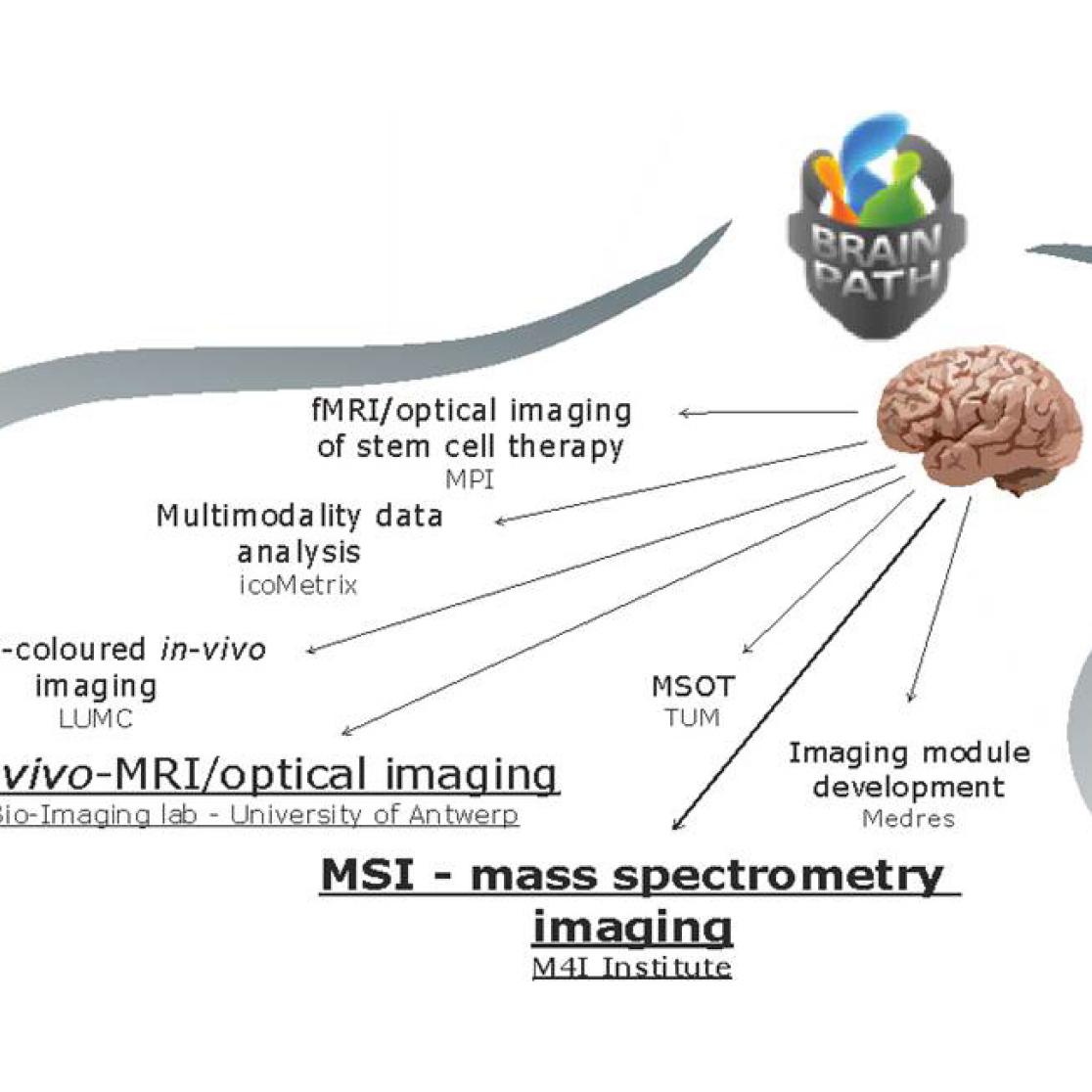
In-vivo imaging
With the concerted application of different high resolution MSI modalities in combination with in-vivo imaging we can provide a global insight into the different molecular structures involved in disease signalling, inflammation and tissue regeneration.
Visit our website Brainpath
Imaging hypoxia and tumor heterogeneity
Partner: Astra-Zeneca
In cancer research new drugs and combinations are tested vigorously in a preclinical environment in order to model the safety and effectiveness of new treatments. If the drug is effective in animal models, then these treatments are taken forward into clinical trials involving human patients.
To generate the preclinical model, usually modified cells from a human cancer are injected into a mouse and grown into a model tumour called a ‘xenograft’. Drugs are administered at carefully controlled doses and quantitative measurements taken of the tumour response. These measurements assume a uniform reaction across the mass of the xenograft. However both in preclinical models and clinical tumours the interior can be highly heterogeneous in both physiological and molecular properties; this is known as intra-tumour heterogeneity. This heterogeneity can drive very different reactions to the same drug within a single tumour. The loss of this detail in preclinical modelling may reduce the translatability of preclinical results into the clinical environment meaning that the effectiveness of new drugs can be misrepresented.
In this project we work with AstraZeneca to investigate molecular profiles that can be used to delineate regions of the intra-tumour heterogeneity. We focus on regions of low oxygen ('hypoxia’) due to leaky blood flow, and dead tissue that forms as a result of chronic hypoxia (‘necrosis’). Both of these factors have been shown to influence the behaviour and response of tumours to drugs in human disease. We use a combination of mass spectrometry imaging, optical imaging, and immunostaining in adjacent tissue sections to extract molecules that distinguish hypoxia and necrosis from viable tissue. For this we have developed a new analysis pipeline that can now be applied to other microenvironmental factors such as blood flow, or immune cell infiltrate. Hypoxia and necrosis - associated profiles are now being investigated in different cancer types to see if they can be consistently used to indicate these microenvironments.
Clinical testing?
The final goal is to integrate these profiles into the preclinical workflow to improve the detail included and the accuracy of the measurements taken to indicate whether a drug should or shouldn’t be taken forwards into clinical testing.
TargetCare: New approaches to joint diseases
Mobility is very important for human well-being, but is seriously impaired by osteoarthritis (OA) and intervertebral disc (IVD) degeneration in many people worldwide. This is mainly due to the degeneration of the cartilaginous tissue of joints.
The TargetCaRe project we are involved in, is part of Horizon 2020 Marie Sklodowska Curie Actions and involves 15 young scientists from 14 partner institutions from 5 different countries all over Europe, aiming to develop treatments for joint diseases by combining advanced drug delivery carriers with dedicated targeting tools and state of the art imaging techniques such as mass spectrometry imaging. The ultimate goal is to prevent further degradation of a damaged OA joint or IVD, as well as to activate the body’s own regenerative capacity to heal damaged and degenerated tissues.
In particular, we will be applying mass spectrometry imaging combined with in vivo imaging to provide proof of principle of the efficacy of new drug delivery systems and to identify cartilage degenerative and regenerative biomarkers. More precisely, TOF-SIMS will be used to study the distribution of the nanocarriers due to the high spatial resolution procured by this technique and MALDI-MSI to analyze the metabolite, protein and lipid regulation after compound administration and treatment with nanoparticles.
Visit our website TargetCare
PRISAR: Innovation in intra-operative diagnostics
PRISAR (pre-clinical intraoperative image guided surgery and postoperative radiotherapy of tumors) is supported by and carried out within the H2020 program RISE funded by the European Commission. The aim of the project is to promote skills development, transfer of knowledge and networking through cooperative development of intraoperative image guided surgery and postoperative radiotherapy of pancreatic tumors by hospital, academic and industrial groups throughout The Netherlands, United Kingdom, France and Germany. The scientific work packages aim to: 1) Develop a near-infrared labelled contrast reagent and tag with optimized affinity and selectivity for newly developed blood vessels; 2) Enhance visual detection of tumor margins by optimizing a Multi-Spectral Optoacoustic Tomography (MSOT) prototype, to detect tissue structures by sound and light) to an unprecedented tissue depth during small animal surgery; 3) Validate the fluorescent construct/antibody for integrin aub3 target combination across the subcellular, cellular, endoscopic and macroscopic levels; and 4) Capture locally applied radio-therapeutics after intraoperative image-guided surgery to demonstrate improved survival rates by improved radionuclide delivery to tumor without increase dose exposure to normal tissue. Additionally, cross-training schedules between 13 groups, transfer of knowledge and oversight of progress to meet deliverables of each work package is managed by Alan Chan of Percuros. Complete information on the PRISAR project H2020-MSCA-RISE grant number 644373-PRISAR.
Termed, molecular pathology, the information contributed to WP2 by M4I-IMS will assist in the ultimate goal of improving pancreatic tumor resection success for patients by providing complementary molecular information about the tumor/non-tumor interface from tissue obtained using image guided surgical techniques being developed in WP1 and WP2. M4I-IMS shares its results and findings not only with the PRISAR consortium, but also with members of the scientific community and general public. Knowledge sharing meetings, contributions to scientific conferences, hosting open days, hosting symposiums and cross trainings are a few examples of how M4I-IMS participates and leads in the skills development, transfer of knowledge and networking within and outside of the PRISAR consortium.
M4I-IMS at Maatricht Univerversity is involved in work package 2 (WP2) in conjunction with Ithera (pre-clinical MSOT) and Mauna Kea Technologies (Cellvizio) with the aim to investigate the tumor/non-tumor interface using mass spectrometry imaging techniques to identify minimum tumor margin distance based on molecular profiles.
Visit our website PRISAR
Imaging Mass Spectrometry in 3D digital pathology
Partners: ITEA, Philips, AMC, PS-MedTech
A strong growth forecast in the digital pathology market for the next five years combined with a decreasing number of qualified pathologists will lead to a tremendous increase in workload in the pathology departments of clinical and pharmaceutical organisations. On top of this there is an urgent need for higher quality diagnostic information enabling more effective and efficient treatments. The ITEA 3DPathology consortium will address these needs by creating a fast, digital, quantitative, spectroscopic and multimodal 3D pathology analysis system.
Imaging Mass Spectrometry will be an integrated part in this analysis system as it is expected to play an important role in future clinical routines for diagnostic and prognostic decision making.
Data analysis
As analytical partner in this ITEA project, M4I is leading the data acquisition work package and will provide multiple 3D imaging mass spectrometry datasets of clinical tissues and participate in the work package for data analysis.
Intra-operative diagnostics for
oncological procedures
Molecular information based surgery - Toward intraoperative MS-based
diagnostics for removal of hepato-pancreato-biliary (HPB) and breast tumors.
Partners: AZM-General Surgery & AZM-Department of Otorhinolaryngology |
Head and Neck Oncology
With more than 3 million new cases and 1.7 million deaths each year, cancer is one of the major health concerns in the modern European society, with continuously increasing incidence. Among the most prevalent forms are hepato-pancreato-biliary (HPB) cancers affecting the liver, pancreas and bile ducts; colorectal cancer (CRC); and breast cancer - the leading type of cancer in women. In most cases, surgery is the only curative treatment possibility, on the condition that the tumor has been diagnosed early enough. The patient, who is facing a life-changing diagnosis, has to undergo an invasive procedure with relatively high risk of recurrence, implying further surgical procedures with a low rate of long-term survival.
Improving the efficiency of surgery is bound to fundamentally improve the prognosis and quality of life for patients with the challenging diagnosis of cancer. The key hypothesis here is that accurate determination of the resection margin between tumor and non-tumor/healthy tissues and thus an improved rate of R0 resections in tumor surgery would reduce the recurrence rates in patients by lowering the risk of residual tumor tissue that remains during the intraoperative process. This would minimize removal and/or damage of healthy tissues as an undesirable effect for the patient and thus would potentially improve patient outcomes and survival rates. In collaboration with the Departments of Surgery and Pathology at MUMC+ we aim to deliver a novel molecular-guided platform for improving ex vivo cancer diagnostics and real-time diagnostics approaches during surgery. The first step will rely in building comprehensive molecular and histopathological classifiers (i.e. e-biobank) for HPB, CRC and breast cancers; which will potentially reveal new candidates for targeted therapies. The second step will be the incorporation of molecular-guided surgery tools for intraoperative MS-based diagnostics.

Identification of cancerous tissue
This innovative approach will present a more precise and accurate identification of cancerous tissue during surgical resection procedures and thus potentially improving the diagnosis and prognosis of cancer patients.
Metabolic profiling of bone during trauma surgery
Regenerative medicine in orthopedic trauma – toward the integration of bone and cartilage metabolic profiling in surgical setting. Partners: AZM-Orthopedics & MERLN
The incidence of fractures in the European population is increasing significantly due to the increasing number of elderly patients at risk. Of all fractures 20% needs to be treated surgically, involving an estimated 900,000 patients. Improvement in operative fracture exposure and fixation techniques has been substantial in recent decades, even for fixation of more difficult osteoporotic fractures in the elderly. It is estimated, however, that in up to 35% in high risk patients, fractures do not heal properly. In addition, at present, prediction of these post-traumatic complications – such as osteoarthritis –at fracture surgery is inaccurate, and not based on bone metabolism and the delayed presentation of complications further compromizes adequate treatment. There is therefore an urgent clinical need of new and innovative approaches to improve surgical procedure and patient’s management.

In close collaboration with the clinical research groups of the orthopedic and trauma surgery departments at azM and MERLN we investigate new MS based surgical tools to study bone and cartilage metabolism. This approach will provide us with an in situ and in real-time diagnostic technique and guide newer therapies aimed at regeneration of the bone and cartilage.
Molecular histology with imaging
MS for colorectal cancer studies
Partner: AZM-Pathology
Colorectal cancer is the third most common cancer worldwide with over a million new cases diagnosed every year. Due to the implementation of CRC screening programs, the number of early stage CRC cases is increasing steadily. However identifying which of these cases are especially high risk, or would benefit from specific treatments such as neoadjuvant therapy (chemotherapy before surgery), is extremely challenging. The tumour node-metastasis staging (TNM) system, which rates the characteristics of the primary tumour, and invasion of nearby lymph nodes and distant metastasis, is the current gold standard, but performs sub-optimally in these early stage cases.
Many other clinico-pathological and molecular characteristics are therefore under investigation for their prognostic and predictive value. These include detailed investigations into tumour characteristics, surrounding lymph nodes, and metastasis, as well as genetic factors such as microsatellite instabilities, DNA methylation, and mutation and expression profiles. Although several show promise, none have yet been implemented clinically. Either the prognostic value is too low, or a standardised definition of the feature is unavailable, or, most commonly, the markers lack appropriate validation and testing in large, non-biased cohorts with proper statistical testing to prove their additional prognostic value in comparison to the current TNM system.
In this collaboration we apply mass spectrometry imaging to analysing the molecular composition of hundreds of CRC patient samples, in combination with full clinical data, staining, genomics, and immunohistochemistry. The molecular profiles will be investigated for potential CRC tumour subtyping.
Analysing the molecular composition
Evidence suggests that several types of CRC cancers exist, each with distinct prognostic characteristics, however a reliable system for distinguishing these has not yet been discovered. Furthermore the full complex data will be carefully analysed to develop a multivariable prediction model of prognosis for various clinical factors including 5 year survival and CRC staging. Further cohorts are available to validate the model, assisting its clinical acceptance, and giving this project a unique opportunity to support individualised treatment and treat high risk CRC cases at an early stage.
Molecular imaging of
osteoarthritic cartilage and regeneration
Partners: AZM-Orthopedics, University Twente, DTL, ZonMW
Osteoarthritis (OA) is the most prevalent form of arthritis. OA affects 80% of the population over the age of 65. It is a complex pathology because diverse factors interact causing the process of deterioration of the cartilage. The current view is that OA is a group of diseases that can be differentiated based on the risk factors and on the pathophysiological mechanisms underlying the joint damage. However, current trials with potential therapies do not distinguish between different subtypes of OA.
Application of MSI in the field of drug delivery also provides insight in the relation between tissue distribution, activity in target tissue and possible local side effects in other tissues, e.g. osteoporotic bone changes, small molecule and tissue characteristics determining penetrance. Local controlled delivery of drugs circumvents side effects of systemic administration while reducing dosing height and frequency. The effectivity of drug delivery systems depends on their release kinetics and dosage. Typically, in the preclinical stage of development, this is measured by determining serum levels or, if available synovial fluid. However, serum levels do not reflect tissue content, as penetration is highly dependent on tissue and drug characteristics. In collaboration with UMCU we aim to reveal the distribution of two anti-inflammatory drugs released from locally injected microspheres in joint tissues, such as cartilage, tendon and bone by MSI. This approach will allow for further fine-tuning of the controlled release-drug combinations and understanding of the principles of controlled release key in the development of efficient drugs. With the support of DTL-associated Technology Hotels and Life Sciences Health funding (ZonMw) we expect to obtain a head start on this novel crossroads between medical biology, drug delivery and molecular imaging.
Mass spectrometry imaging (MSI) has become a powerful method for tissue-based disease classification
The power of MSI resides in the ability to detect lipids, proteins, drugs, metabolites and other compounds directly from tissue whilst preserving the information about their spatial localization. Preliminary work of our group has classified OA and healthy groups based on the peptidomic and lipidomic profiles of human cartilage and synovium.
In collaboration with the Orthopedic Surgery department at MUMC+ and the division of Rheumatology at azM we aim to identify different OA phenotypes by MSI and use this novel high throughput strategy to reveal new markers for a personalized medicine.
Intra-tumor heterogeneity
and personalized medicine
Partners: Helmholtz Zentrum Muenchen, Munich, Germany, Fondazione della Scienza ONLUS, Pisa, Italy, West German Cancer Center, Essen, Germany, Klinikum rechts der Isar, Munich, Germany, Institute of Genetics and Molecular and Cellular Biology, Strasbourg, France.
In many tumors, including esophageal and esophagogastric junction adenocarcinomas (EAC), intra-tumor molecular heterogeneity is closely linked to tumor characteristics that are responsible for a poor prognosis of the patients such as therapy resistance, early metastasis and tumor recurrence. Hence there is a strong need to identify clinically-relevant tumor subpopulations.
In this study, we will use Imaging Mass Spectrometry to investigate intra-tumor molecular heterogeneity in EAC with the aim to identify robust molecular classifiers which help to direct therapy based on an analysis of sequential tumor material from the MEMORI trial (Metabolic and Molecular Response Evaluation for Therapy Individualization in esophageal and esophagogastric junction adenocarcinomas, EUDRA-CT Number 2014-000860-16). In this clinical trial, 14 days before and after induction of neoadjuvant chemotherapy tumor response is evaluated using PET and sequential tumor biopsies obtained. This setting gives the unique opportunity to study the evolution of intra-tumor heterogeneity of EAC in the context of therapy response through a molecular examination of the tissue specimens before, under and after neoadjuvant therapy.
Multiplex molecular information
Imaging Mass Spectrometry has the unique capability to decipher intra-tumor heterogeneity as it allows reading out multiplex molecular information of tissues in a spatially-resolved way without any label.
Unraveling the complexity of troponins in human heart
by imaging mass spectrometry
Partner: azM Clinical Diagnostic Laboratory
Troponin is a unique and heart specific protein (troponin T or I) and is released into the bloodstream after an infarction. Using the recently introduced high sensitive troponin immunoassays it is also possible to detect troponin in healthy individuals. Increased troponin concentrations are found in diseases like heart failure, myocarditis, renal failure and also endurance exercise. This complicates the anamneses of the typical and atypical chest discomfort to a cardiac cause.
Here, we target the investigation of the release of cardiac troponin by cardiomyocytes, using clinical diagnostic tests and imaging mass spectrometry. Focusing on understanding the local molecular changes in myocardial dysfunction on one hand and unraveling metabolic patterns and pathways of troponin release on the other hand. Native mass spectrometry can be used as well to investigate the conformation of the troponin complex with and without the effect of Ca2+ and/or implicated enzymes. All together this can give insight in the time dependent degradation pattern of troponin, a possible correlation with the stability of atherosclerotic plaques or the diagnostic distinction between the acute and chronic elevation of troponin concentrations.
Unraveling metabolic patterns
Focusing on understanding the local molecular changes in myocardial dysfunction on one hand and unraveling metabolic patterns and pathways of troponin release on the other hand.
Forensic hair testing
Hair testing is a powerful tool routinely used for the detection of drugs of abuse in toxicology and forensic applications. The analysis of hair is highly advantageous as it can provide prolonged detectability versus that in biological fluids and chronological information about drug intake based on the average growth of hair. However, current methodology routinely involves complex and time-consuming sample preparation followed by gas or liquid chromatography coupled with mass spectrometry. This project has investigated the use of mass spectrometry imaging techniques to visualise the distribution of drugs of abuse (such as cocaine) in single human hair samples, these techniques offer the ability to provide more accurate and visual chronological information on drug intake (hours to days) over current techniques which only provides values on a monthly basis.

These techniques have also been used to investigate the effects of current forensic decontamination protocols and cosmetic treatment on the analysis of drug user hair samples.
Quantitative molecular imaging
in pharmaceutical sciences
In the pharmaceutical industry Mass Spectrometry (MS) is an integral analytical tool used throughout novel drug research, e.g. in discovery, DMPK, in pre-clinical and clinical toxicology, impurity profiling, stability testing, efficacy testing etc. The use of Imaging MS though us still relative new and visualizing spatial distribution of an active drug and associated metabolites or complete affected metabolic pathway offers still a lot of challenges. On the other hand spatial information of new (and existing) drugs and associated metabolites in tissues will improve mechanistic understanding and clinical consequences of pre-clinical safety concerns and will direct better decision making in further drug development. Besides, local spatial and quantitative information will allow PK/PD assessment at the tissue level as well as target/pathway engagement. In combination with MS based (metabol)omics it represents a highly promising approach in mechanism based safety evaluation as well as (novel) target identification/validation and will prove to be extremely valuable across all therapeutic areas.
Severe challenges
As mentioned though there are still severe challenges, like e.g. attainable spatial resolution of low to sub um level, reliable quantitative information and improved sensitivity of IMS. Therefore, we have shaped three separate projects to address these challenges and at the same time apply the developed technology in relevant projects and real-life examples.
Proteins are the most common molecules found in cells. In fact, they constitute more of a cell’s dry matter than lipids, carbohydrates and all other molecules combined. Proteins come in a huge variety of forms and perform a wide range of functions. Examples of proteins include enzymes, antibodies and some hormones, which help to speed up chemical reactions, defend against diseases and regulate the activity of cells. Their function depends on their structure, chemical and physical properties as well as their abundance in specific tissues.
Selection of Proteins
In this project students will investigate structure, function, tissue specificity and disease association for selected proteins. Proteins can be selected based on known disease associations or experimental results. We also encourage students to come up with their own suggestions.
Analysis
Following aspects of the proteins will be investigated:
- Protein Structure: primary, secondary, tertiary and quaternary.
- Effects of genetic variations on the protein structure
- Splice variants
- Protein Domain conservation
- Protein Function or functional predicted function
- Tissue abundance (Tissue specificity)
- Disease associations
Link to other projects
This project is part of a multi-layered system biology approach to study human genes, proteins and their functions. Students will investigate the molecules of interest on the DNA level (“Are you being conserved: sequence analysis of candidate genes identified in hereditary disorders”, ID:3708), the protein level (“Proteins under investigation”, ID:3709) or the functional interactome level (“Connecting the dots: constructing interaction networks of candidate disease genes ”, ID:3710).
| Apply for this internship |
|---|
Contact
Name: Martina Kutmon
Phone: +31 43 388 1123
Email: martina.kutmon@maastrichtuniversity.nl
Abroad
Placement outside the Netherlands: No
Erasmus subsidy: No
Mastery of languages: English
Ethical review
MERC (medical): Not applicable
AEC/DCC (animal experiment: Not applicable
ECP (psychology): Not applicable
Additional
Possible to do internship in English: Yes
Type of internship
Thesis on the basis of an existing data set
Thesis based on own data collection
First supervisor
Name: Gökhan Ertaylan
Phone: +31 43 388 2913
Email:gokhan.ertaylan@maastrichtuniversity.nl
Department: Maastricht Centre for Systems Biology
To build these profiles, several large molecular datasets from various deeply phenotyped cohort studies are available. These datasets are generated from whole blood, which immediately complicates matters: blood contains a range of cell types, each with a unique molecular profile. Additionally, the relative amounts of each cell type vary from person to person. Although this information is hidden, it is not very well hidden: it can be derived numerically from the data using specialized algorithms. Within this project we would be exploring several such algorithms that have recently been published that are capable of performing the task at hand.
The results will form the basis for a network based analysis: cell type specific, disease related changes in regulation of specific biological networks.
Type of Research
Programme
-
Systems Biology (FSE)
-
Maastricht Science Programme (FSE)
-
Data Science and Knowledge Engineering (FSE)
-
Biomedical Sciences (FHML)
-
Bachelor and Master level
Contact person/supervisor
- Name: Michiel Adriaens, Assistant professor
- Department: Maastricht Centre for Systems Biology
To apply for ethical review, please fill out the application form and send it to the secretary. For help with filling out the application form, please consult these tips and tricks.
Please note that any substantial changes in your research design after ethical approval require a renewed review by ERCIC. To apply for an amendment to your original protocol, please send an email to the secretary describing the changes you have made in the email, and highlighting the changes in the updated application form plus annexes (if applicable).
ERCIC members
Valentina Mazzucato (interim chair)
Greg Stapleton (secretary)
Prof. Frank Corvers (SBE)
Dr. Robert Gianni (FASoS)
Prof. Remco Havermans (FSE)
Dr. Jona Linde (SBE)
Prof. Niels Philipsen (Law)
Dr. Soetkin Verhaegen (FASoS)
Prof. Lisa Waddington (Law)
Prof. Gerhard Weiss (FSE)
ERCIC deadlines 2024/2025
ERCIC meets every 4 weeks during this academic year to consider applications for ethical review:
| Submission deadline (12:00 midday) |
|---|
| 28 August 2024 |
| 25 September 2024 |
| 23 October 2024 |
| 20 November 2024 |
| 8 January 2025 |
| 5 February 2025 |
| 26 February 2025 |
| 2 April 2025 |
| 30 April 2025 |
| 26 May 2025 |
| 26 June 2025 |
ERCIC documents
News on ERCIC
Procedure
Since September the procedure for digital data storage devices like USB sticks, DVD’s, but also mp3 players and telephones has changed. These items are forwarded to ICTS after one week.
When devices are not collected they are eventually destroyed (to protect the privacy of the owner) or delivered to the Lost and Found department at the Maastricht municipality (for high value items).
You can still collect your digital storage device for one month at the ICTS Front Office at Grote Looiersstraat 17. We will check our stock of items found if you call us and describe the device, the date and location where you lost it and what’s on the device. When you come to collect it we’ll ask you to show that the information described is actually stored on the device (this will be done on your own laptop when needed).