M4I Facilities
Both divisions of M4I are equipped with cutting-edge scientific equipment.
Division of Nanoscopy Infrastructure
Cryo-electron microscopy is the only way to study cellular processes close to the in vivo situation. In order to do so, we have implemented a full workflow, starting with life cell imaging of cells or tissues followed by rapid cryo-fixation to preserve the structure. Cryo-light and Cryo-electron microscopy will resolve the 3D structure at nm resolution by means of electron tomography.
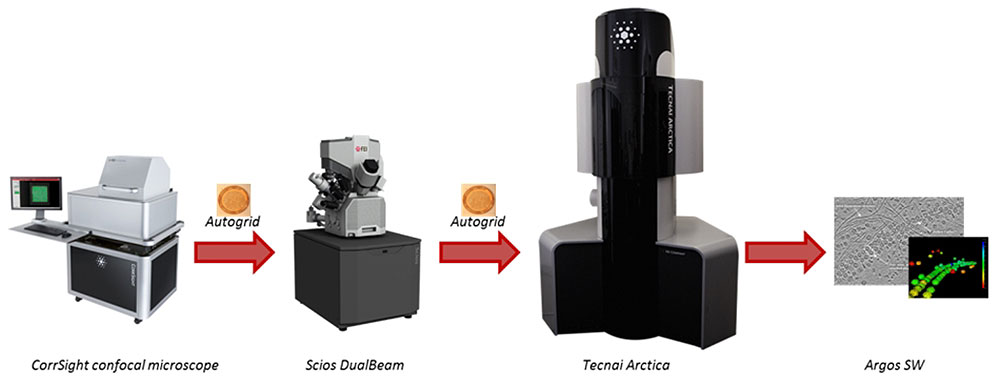
Our full cryo-correlative workflow for ‘Selected volume tomography’
More equipment





Ultramicrotome Leica EM UC7
The Ultramicrotome Leica EM UC7 provides easy preparation of semi- and ultrathin sections as well as perfect, smooth surfaces of biological and industrial samples for TEM, SEM, AFM and LM examination.
Cryochamber Leica EM FC7
The Leica EM UC7 can be transformed into a Cryoultramicrotome by mounting the Cryochamber Leica EM FC7 to prepare cryo-sections ( -15° to -185°C) for TEM, SEM, AFM and LM.
The Leica EM AFS2
The Leica EM AFS2 performs freeze substitution and progressive lowering of temperature (PLT) techniques and allows low temperature embedding and polymerization of resins.
The Leica EM FSP (freeze substitution processor), an automatic reagent handling system combined with the Leica EM AFS2, dispenses reagents for both freeze substitution and PLT applications. The LED illumination from within the chamber and the attached stereomicroscope for viewing and positioning of samples ensures ease of use.
Leica TCS SP8 CARS confocal microscope
For imaging with the Leica TCS SP8 CARS confocal microscope, specimen do not have to be stained. The Coherent anti-Stokes Raman Scattering (CARS) technology involves two laser beams to visualize the vibrational contrast of molecules in specimens. The specimen remains almost unaffected.
Combinie the iCorr fluorescence light microscope module and the FEI Tecnai electron microscope
Combining the iCorr fluorescence light microscope module and the FEI Tecnai electron microscope into a single, harmonized instrument affords users a streamlined approach to correlative microscopy. This workflow requires no sample transfer, allows capturing fluorescence data as well as high resolution electron microscopy data and correlates that data in minutes.
Watch the video for more information.
In addition to the CorrSight, Scios DualBeam, Tecnai Arctica, the Division of Nanoscopy also has the following equipment, which can be seen in the slideshow:
- Ultramicrotome Leica EM UC7
- Cryochamber Leica EM FC7
- Leica EM AFS2
- Leica TCS SP8 CARS confocal microscope
- Combined iCorr fluorescence light microscope module and the FEI Tecnai electron microscope
Division of Imaging Mass Spectrometry Infrastructure
M4I has a unique infrastructure for state-of-the-art mass spectrometry based on molecular imaging of complex surfaces. Complementary molecular imaging instruments allow the determination of the spatial distribution of intact, unlabeled biomolecules from cellular and tissue surfaces with spatial resolutions ranging from 200 nm to 200 micrometer.
The facility is equipped with a variety of sample handling and automated preparation tools that allow the usage of controlled and reproducible protocols. The different desorption and ionization methods available allow the analysis of a variety of complex surfaces, such as semiconductors, paint and coatings, polymers, single cells and histological tissue sections. In addition, various proteomics technologies are used to investigate the structure-function relation of biomolecules in their native environment.

The M4I Division of Imaging Mass Spectrometry is equipped with:
- 9.4T MALDI/ESI Fourier Transform Ion Cyclotron Resonance imaging mass spectrometer
- 7T LTQ-FTICR system for atmospheric ionisation and imaging
- LAESI DP-1000 ambient imaging system of Protea Biosciences
- Bruker Ultraflex III ToF/ToF MALDI molecular imager
- Physical Electronics TRIFT II high resolution secondary ion mass spectrometry (SIMS) ToF-MS system
- TRIFT II based AMOLF mass microscope for direct ion imaging with MALDI and C60-SIMS
- Waters MALDI/ESI Synapt G2Si system for high resolutionmolecular imaging
- Waters MALDI Synapt HDMS system for conformational molecular imaging
- fully equipped sample preparation facility, including cryosectioning, staining, analytical microscopes, matrix deposition and coating devices
- Advion Triversa Nanomate system
- Nano-HPLC systems
- spotting robot for LC-MALDI
- virtual laboratory based image processing and analysis tools, including tissue classification and correlation software and protein data basing tools
- large scale data storage and cluster computing facilities
Capabilities
Fundamental Cancer Research in the Netherlands
FTICR-MS imaging
Researchers of the M4I Division of Imaging Mass Spectrometry pursue analytical methods to determine the identity and location of biomolecules down to the subcellular (micrometer) level. Available mass spectrometric imaging methods either compromise localization accuracy or identification accuracy in their analysis of surface biomolecules.
MALDI-FTICR-MS is developed for the spatially resolved mass analysis of rat brain tissue with the aim to optimize protein identification by the high mass accuracy and online MS/MS capabilities of the technique. Mass accuracies down to 1 ppm were obtained in the direct MALDI-analysis of the tissue together with a spatial resolution of 200 micrometer. The spatial distributions of biomolecules differing in mass by less than 0.1 Da could be resolved and are shown to differ significantly. Online MS/MS analysis of selected ions was employed for endogenous peptide identification.

Imaging detectors
Understanding the structure and the composition of biological systems requires imaging systems that push the limits of spatial resolution and molecular sensitivity. This project aims to implement new imaging detector systems based on the Medipix/Timepix chip on our ion microscope mass spectrometer for applications in molecular histology. This work is conducted in collaboration with the detector R&D group of Nikhef in the framework of a joint STW project.
Instrumentation
Several technical challenges are related to the in-vacuum mounting of the Medipix/Timepix pixel detectors. Technical developments related to the in-vacuum mounting of the chips as, for instance, a vacuum-compatible chip carrier and active temperature control of the chips will be presented here soon.
Outlook
The performance of the current Medipix/Timepix detector is limited by the counter depth and the single-stop pixels. As a future perspective, we envisage a successor Timepix chip that incorporates 100 ps TDC bins, a 1 ms total measurement interval and multi- (double-) stop pixels. Standard TOF-MS detection systems are already shown to be outperformed by several unique Timepix system capabilities. These are the combination of S/N, multiplexed detection, dynamic range and the simultaneous detection of position- and time-information by a single detector system.
The combination of this MCP/Timepix detection system and an ion microscope constitutes an extremely powerful MSI instrument for high dynamic range, high mass range and high detector homogeneity studies of intact proteins of bio-molecular relevance.

Imaging detectors: Fast, High Resolution Mass Spectrometry Imaging Using a Medipix Pixelated Detector
(Top) Schematic representation of the setup elements such as the sample, surface probe, ion microscope, electrostatic blanker, MCP and Medipix2 detector. (Bottom) Potential diagram of the experimental setup. The surface probe desorbs and ionizes the sample surface. The ionized surface particles are electrostatically extracted and guided through the mass spectrometer by the ion optics. Particular m/z species can be selected from the full-ion load with the electrostatic blanker. The (mass-selected) ion load is detected by the chevron MCP stack and Medipix2 detector.
In mass spectrometry imaging, spatial resolution is pushed to its limits with the use of ion microscope mass spectrometric imaging systems. An ion microscope magnifies and then projects the original spatial distribution of ions from a sample surface onto a position-sensitive detector, while retaining time-of-flight mass separation capabilities. Here, a new type of position-sensitive detector based on a chevron microchannel plate stack in combination with a 512 × 512 Medipix pixel detector is coupled to an ion microscope.
Advantages of CMOS pixel detectors in mass spectrometry imaging are the ease of data acquisition and data processing (direct image acquisition), the high spatial resolution (possibly enhanced by centroiding algorithms), the single-particle counting mode (noise-free particle counting as opposed to charge integration in CCD-based detectors) and the capability of some detector generations to take time-resolved images (Timepix).
Given these attractive CMOS detector features, we have demonstrated the technical, in-vacuum implementation and capabilities of this novel type of charged-particle detector –a chevron MCP stack in combination with a Medipix2 imaging readout chip- for microscope mode MSI.
This detection system has 55 × 55 µm2 detector pixels and an ion optical magnification of a factor 85 (SIMS) and 42 (LDI) such that an individual pixel probes an area of 650 × 650 nm2 (SIMS) and 1.31 × 1.31 µm2 (LDI) on the sample surface. The detector is capable of imaging organic material with ≤ 6 µm spatial resolving power by positive mode SIMS and with 8- 10 µm spatial resolving power using LDI mass spectrometry imaging.
A detailed evaluation of key performance criteria such as spatial resolution, acquisition speed and data handling is performed during a collaborative study with the detector R&D group of Dr. Jan Visser at FOM-NIKHEF.

Imaging detectors: High Dynamic Range Bio-Molecular Ion Microscopy with the Timepix Detector
A schematic representation of the experimental setup. An MCP-Timepix detector assembly is coupled to a commercial ion microscope. Mass spectra, total and mass-selected ion images are generated from one data set without the need for mass selection and repetitive measurements. The decoupled MCP signal provides a single signal that integrates all events and is acquired by a multi-stop TDC and an ADC.
Highly parallel, active pixel detectors enable novel detection capabilities for large bio-molecules in time-of-flight (TOF) based mass spectrometry imaging (MSI). In our detector development project, a 512 × 512 pixel Timepix assembly combined with chevron microchannel plates (MCP) captures time-resolved images of several m/z species in a single measurement. The use of a MCP-Timepix assembly delivers an increased dynamic range of several orders of magnitude. The Timepix returns defined mass spectra already at sub-saturation MCP gains, which prolongs the MCP lifetime and allows the gain to be optimized for image quality. The Timepix peak resolution is only limited by the resolution of the in-pixel measurement clock. Oligomers of the protein ubiquitin were measured up to 78 kDa.

Ion Mobility Imaging
In collaboration with Waters and the Netherlands Proteomics Centre a new research instrument was added to the molecular imaging facility at AMOLF. Since January 2009 a SYNAPT HDMS is employed to separate biomolecular ions based on their gas-phase collision cross-section using ion mobility spectrometry. The instrument has been transferred from AMOLF to Maastricht University at the end of 2014. The research capabilities of M4I have been recently expanded with an additional state-of-the-art MALDI Synapt G2Si for ion mobility based imaging MS through an investment by Enabling Technologies.
The SYNAPT systems are the only commercial instruments to incorporate high efficiency ion mobility separation in a MALDI-ToF mass spectrometer giving the scientists maximum analytical power for the discovery of research challenges. Molecular images are generated from biomedical tissue sections that contain information on the molecular weight of the surface biomolecules, their ion mobility properties and their spatial distribution. The coupling of ion mobility separation with imaging MS realizes innovative possibilities to separate nominally isobaric species prior to mass analysis.
This will result in the visualization of new spatial features directly from biomedical tissue and help in understanding the underlying molecular causes of various diseases. The first application of this innovative instrument is foreseen in the breast cancer research effort in collaboration with Johns Hopkins Medical School in Baltimore.

e-biobanking
The M4I Division of Imaging Mass Spectrometry participates in the BSIK funded COMMIT project for virtual tissue biobanking. This project aims to establish an environment to process and evaluate large datasets originating from molecular histology using imaging mass spectrometry. A concise statistical analysis for the extraction of biomarker patterns from the molecular images is targeted to enable an innovative diagnosis approach in a biomedical imaging environment. Protocols will be established and employed to generated detailed mass spectrometry based molecular image datasets. The extracted molecular images together with their corresponding descriptive metadata will be stored in the Netherlands Virtual Tissue Bank.
In close collaboration with TI-COAST, the research group will establish a workflow based on a knowledge management system that will build upon the KnowEx system developed in VL-e. Distributed processing of the large image datasets is considered using the IBIS cockpit system developed at the VU computer science group. The applications will be tested on breast cancer sections collected from the medical partners in this project. A significant amount of data acquisition of a breast cancer tissue collection will be generated. The virtual tissue bank will be filled with the molecular mass spectrometric images generated in the framework of this program from these sections to act as a reference database for medical professionals. The tissue microarray data of these diseases will be linked with the histological images when available.

Imaging Proteomics
On-tissue enzymatic digestions for direct protein identification
A participation in the Netherlands Proteomics Center - NPC.
Mass spectrometry is rapidly maturing as a powerful tool for proteomics applications as new sample-preparation approaches, instrumentation developments and bioinformatics tools for data analysis become increasingly available. No other technology can now match both the throughput and the molecular information content that mass spectrometry provides. Instrument manufacturers have made a tremendous effort in the last decade and have come up with innovative instrumentation which continuously allows more speed, sensitivity and specificity for standard high-throughput proteomics.
As a partner in the Netherlands Proteomics Center (NPC II), the M4I Division of Imaging Mass Spectrometry aims to progress beyond the standard mass spectrometry based analysis. To this end, a platform for spatially resolved mass spectrometric proteomics imaging was deployed, which will be further developed and embedded in the NPC. Imaging mass spectrometry is developed and combined with new proteomics tools to improve local peptide and protein identification directly from unstained and unlabeled thin tissue sections. New protocols for local tissue digestion are under study where tandem MS is used to improve the protein identification capabilities during imaging MS.
The strategies will be combined into a new approach for 3-dimensional MS based imaging of small xenografted tumors and organs. The rendered three-dimensional MS will be evaluated and compared with high resolution MRI 3D images rendering a new tool for the visualization of molecular anatomy.

Molecular histology: Lipid cartography of mouse atherosclerotic plaques by ToF-SIMS imaging mass spectrometry
A collaboration between the M4I Division of Imaging Mass Spectrometry and the Experimental Vascular Pathology group of Erik Biessen at Maastricht University.
The nature and distribution of lipids in atherosclerotic lesions from mice carotid arteries is studied with complementary molecular imaging technique. The lipid content of atherosclerotic plaques determines their mechanical stability and highly thrombogenic lipids such as lysophosphatidic acids (LPA) can be localized by IMS within lesions.
Spatial resolution down to cellular level using Secondary Ion Mass Spectrometry (SIMS) was performed in order to identify, according to their specific composition, cellular layers of different histological type: phosphate and phosphocholine in vascular smooth muscle cells, cholesterol and LPA in necrotic core. This prima example of translational research in molecular medicine illustrates how innovations from physics can be directly beneficial to biomedical research.

Molecular histology: New sample preparation protocols for complementary molecular imaging of brain tissue
A collaboration between the M4I Division of Imaging Mass Spectrometry and the Neuro-Oncology group of Theo Luider and pathologist Max Kros at the Erasmus Medical Centre in Rotterdam.
Different complementary imaging mass spectrometry methods, i.e. matrix-assisted laser desorption/ionization (MALDI) and secondary ion mass spectrometry (SIMS) imaging for direct tissue analysis, can be applied on exactly the same tissue sample. This allows the identification of small molecules, peptides and proteins present on the same sample surface. Sample preparation is crucial to obtain high quality, reliable and reproducible complementary molecular images. It is essential to optimize the conditions for each step in the sample preparation protocol, ranging from sample collection and storage to surface modification. Researchers of the M4I Division of Imaging Mass Spectrometry have investigated the importance of correct sample treatment in case of MALDI and SIMS imaging experiments and describe the experimental requirements for optimal sample preparation.

Molecular histology: Molecules at the interface of biology and polymers
The image shows an overlay of an optical image with a mass spectrometric image measured using SIMS (secondary ion mass spectrometry). The colours in the overlay indicate the localization of the characteristic peaks belonging to cholesterol (red), polyethyleneglycol (green) and choline (blue). Thus polymer and tissue can be localized using their molecular profiles.
Imaging mass spectrometry (IMS) has made a big impact in the life sciences, giving localized chemical information in tissue samples. Although technical developments have mainly focussed on biological systems, they can very well be applied on combined synthetic/biological systems. IMS could be a very valuable tool for the identification of foreign-body reaction and biodegradation at a molecular level in a drug delivery system. Polymer implants were studied in a rat kidney model. Complementary IMS techniques can contribute in the understanding of the properties of polymers in vivo. A more thorough understanding of these properties opens up possibilities for novel applications of polymers for drug delivery or other medical use.

Molecular histology: Hypoxia driven signaling pathways in breast cancer
An NIH funded collaboration between the M4I Division of Imaging Mass Spectrometry and Dr. Kristine Glunde at the Radiology department of Johns Hopkins Medical School in Baltimore, USA.
In collaboration with the Johns Hopkins Medical school in Baltimore, USA, the M4I Division of Imaging Mass Spectrometry studies hypoxia-driven molecular signaling pathways in breast cancer. Using various molecular imaging techniques, both in vivo and histological, molecular distributions in tumors, that have grown from human breast cancer cell lines, are mapped two and three-dimensionally.
For this research, which will last four years starting January 2009, the researchers will use and further develop the high resolution mas spectrometry based molecular imaging. Dr. Kristine Glunde and her group will label selected proteins with GFP, whereas Ron Heeren's group will map signal molecules with the same special distribution. KnowEx, the platform for transfer of knowledge, designed at AMOLF for the "Virtual laboratory for e-science", facilitates the bringing together of the various molecular and anatomical image modalities (MRI, Computer Tomography, confocal imaging and MS imaging). This way the researchers hope to visualize for the first time the molecular players that regulate hypoxia-driven processes in breast cancer cells. At the same time the molecular distributions revealed will be used to perform a rapid classification of the cell types within the tumor.

Molecular histology: Pinpointing pharmaceutical metabolism: "Google Rat"
A key question in pharmaceutical research focuses on the metabolic pathways. Pharmaceutical research employs imaging MS complementary to whole body autoradiography in both research and development phase of new drugs. MS imaging is particularly interesting as it has the potential to image simultaneously the drugs and their metabolites. MS imaging offers the possibility to not only examine and track drug metabolism in whole animal sections, but can also follow polymer substrate degradation and drug release. With a number of pharmaceutical industries the M4I Division of Imaging Mass Spectrometry is developing new imaging protocols for drug metabolic imaging. A protocol called 'Google Rat' allows the researchers to examine molecular distributions in whole body tissue sections and zoom down to the single cell level on the same sample.

- Mass Microscopy
- Fundamental Cancer Research in the Netherlands
- FTICR-MS imaging
- Imaging detectors
- Imaging detectors: Fast, High Resolution Mass Spectrometry Imaging Using a Medipix Pixelated Detector
- Imaging detectors: High Dynamic Range Bio-Molecular Ion Microscopy with the Timepix Detector
- Ion Mobility Imaging
- e-biobanking
- Imaging Proteomics
- Molecular histology: Lipid cartography of mouse atherosclerotic plaques by ToF-SIMS imaging mass spectrometry
- Molecular histology: New sample preparation protocols for complementary molecular imaging of brain tissue
- Molecular histology: Molecules at the interface of biology and polymers
- Molecular histology: Hypoxia driven signaling pathways in breast cancer
- Molecular histology: Pinpointing pharmaceutical metabolism: "Google Rat"
